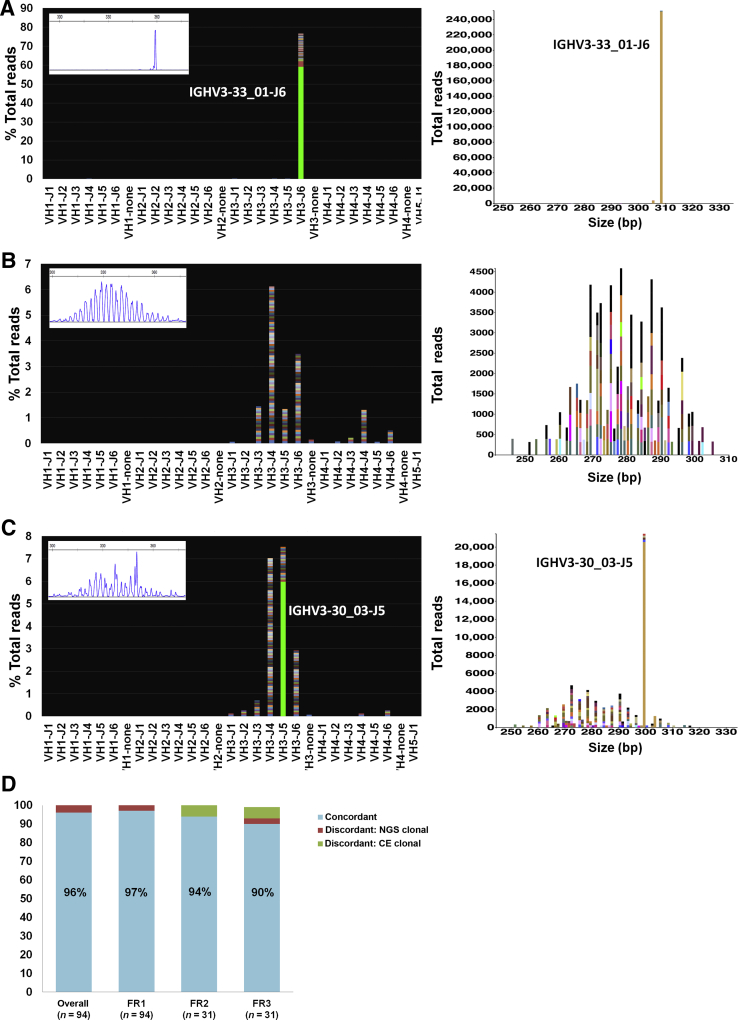
Figure 1.
Representative examples from the validation set. A: Clonal case (Sample 16, diffuse large B-cell lymphoma, formalin-fixed, paraffin-embedded tissue; 330,591 total reads). Left panel: Snapshot of the V-J sequence frequency graph showing the distribution of reads stratified by sequence and family. Most reads are restricted to a rearrangement using V3-33 and J6 family and share an identical sequence, as exemplified by the solid green of the bar, whereas reads with slightly different sequences are stacked on top in various colors. Inset: Corresponding capillary electrophoresis (CE) results (FR1 BIOMED-2 primers). Right panel: Sequences plotted by the Memorial Sloan Kettering (MSK) LymphoClone viewer graphically display the clonal reads based on size and V-J use. B: Polyclonal case (Sample 63, no disease, BM aspirate; 470,767 total reads). Left panel: Snapshot of the V-J sequence frequency graph shows multiple, multicolored bars with a variable distribution of VJ use. Each colored band on a bar denotes a different sequence. Inset: Corresponding CE results (FR1 BIOMED-2 primers). Right panel: MSK LymphoClone viewer shows multicolored bars arranged by size and VJ use in a gaussian distribution typical of a polyclonal pattern. C: Discrepant case (Sample 48, plasma cell neoplasm involving BM, 4% by aspirate differential count; 353,964 total reads). Left panel: V-J sequence frequency graph shows a dominant clonal sequence (solid green bar) within a polyclonal background (multicolored bars). Inset: Corresponding CE results (FR1 primers) show a polyclonal pattern with a slight prominent peak at approximately 350 bp without a definitive clonal Ig heavy chain (IGH) rearrangement. Right panel: Corresponding MSK LymphoClone viewer plot shows a solid yellow bar depicting the clone within a polyclonal background. D: Validation accuracy study for the FR1, FR2, and FR3 primer sets. The overall concordance for a clonality call based on all primer sets targeting IGH was 96%, with superior clonal detection using next-generation sequencing (NGS). Results stratified by individual primer sets show higher discordance rate for FR3 primers.