Figure 3.
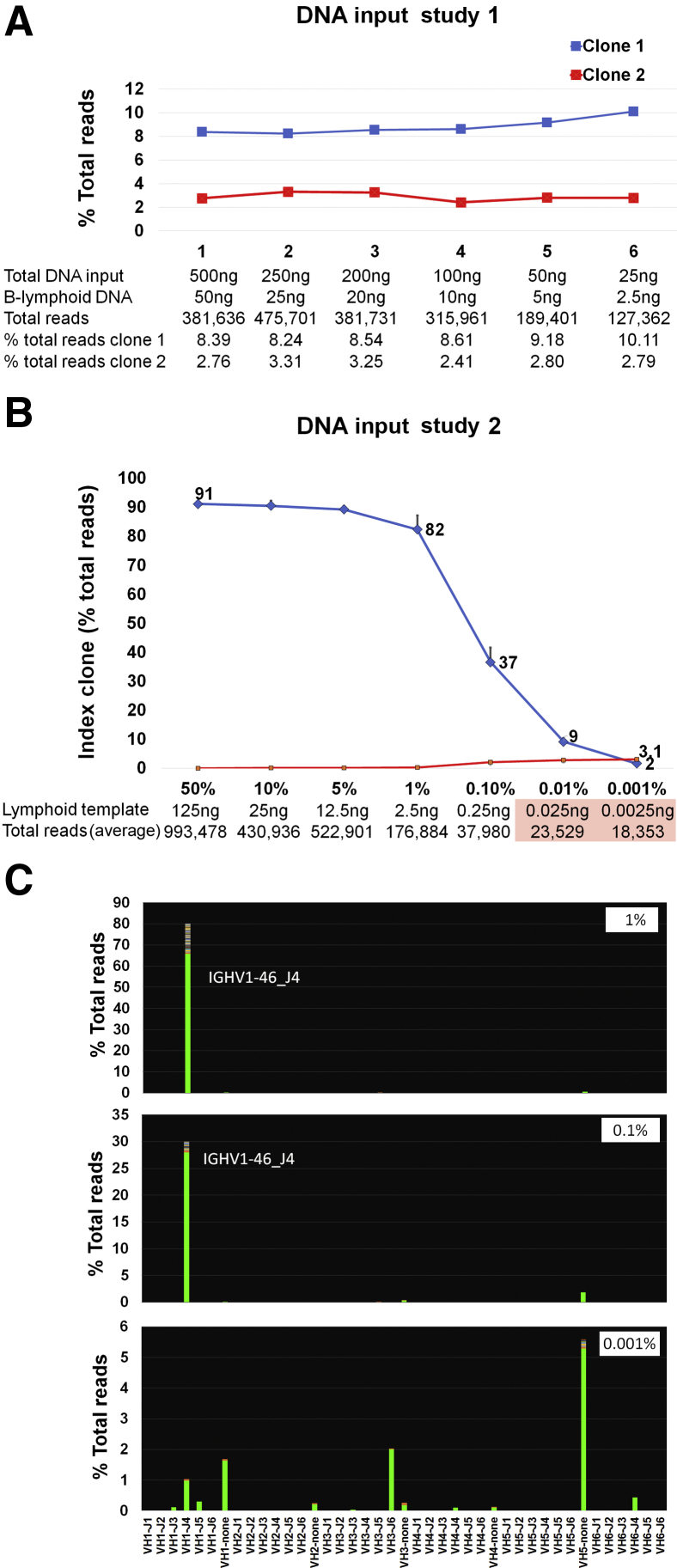
DNA input studies. A: DNA input study using decreasing amounts of total DNA from a patient sample containing approximately 10% B-lymphoid cells. Total DNA input and estimated B-cell–derived DNA detailed at each level. Decreasing the total DNA input from 500 to 25 ng has a moderate effect on the total number of reads but no significant impact on clone detection. B: DNA input study using decreasing amounts of clonal lymphoid DNA into epithelial DNA. Total DNA input is fixed at 250 ng. Graph shows the level of detection of the index clone in a dilution series with sequentially decreasing amounts of clonal lymphoid DNA diluted into epithelial DNA (four replicate reactions performed for each dilution). On the x axis, seven dilutions are shown (50%, 10%, 5%, 1%, 0.1%, 0.01%, and 0.001% lymphoid component in a total DNA input of 250 ng). Details of the estimated lymphoid component at each level and total number of reads generated are seen below. In the undiluted characterized lymphoid sample, the clonal sequence encompassed 90% of the total rearranged Ig heavy chain reads. This clone, and its relative proportion, remained constant down to the 12.5-ng input. Further decreases in the lymphoid DNA input lead to a sequential decrease in the overall proportion of the index clone and preferential amplification of nonspecific (pseudoclonal) sequences. Further decreases in the lymphoid DNA input lead to a sequential decrease in the overall proportion of the index clone and preferential amplification of nonspecific (pseudoclonal) sequences, ultimately leading to a failure pattern as total reads decrease below 30,000 (pink box). C: Representative views of the V-J sequence frequency graphs at 1%, 0.1%, and 0.001%. The 0.001% dilution shows a failure pattern, with multiple bars depicting nonspecific sequences with no J segment; the index clone is not detected.