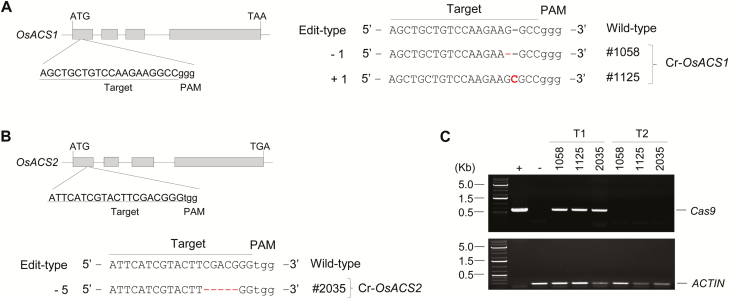
Fig. 3.
CRISPR/Cas9-based mutagenesis on OsACS1 or OsACS2. (A, B) Schematic gene structure of OsACS1 (A) or OsACS2 (B) and detection of targeted mutation(s) (deletion or insertion) in the OsACS1 (A) or OsACS2 (B) locus. The target site nucleotides are shown in upper case letters and the protospacer adjacent motif (PAM) is indicated as lower case letters. ‘–’ and ‘+’ indicate either the deletion or insertion of nucleotide(s), respectively. (C) PCR-based identification of Cas-free OsACS1 (Cr-OsACS1) and OsACS2 (Cr-OsACS2) mutant plants. The gel image shows the presence or absence of Cas in the genomic DNA of T1 and T2 lines of Cr-OsACS1 and Cr-OsACS2. ‘+’ indicates a positive plasmid control used in this study containing cloned Cas9. ‘–’ indicates a negative control genomic DNA isolated from WT rice.