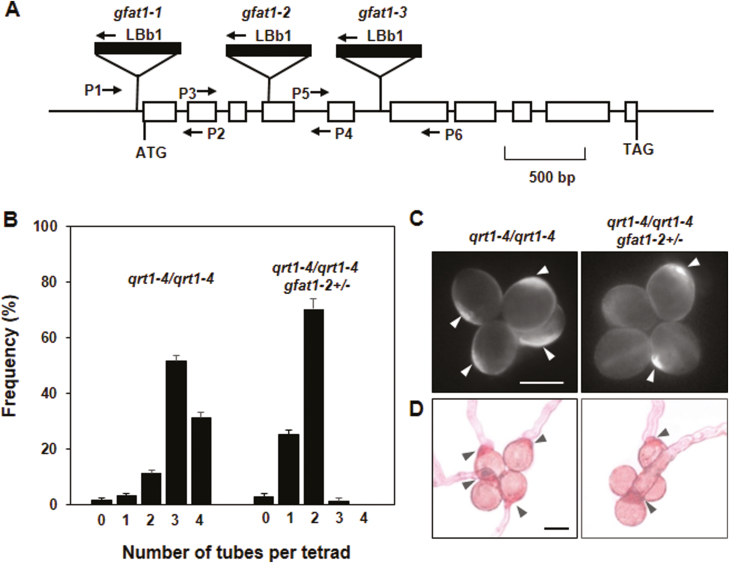
Fig. 2.
gfat1-2 pollen is defective in the polar deposition of cell wall materials. (A) Schematic representation of AtGFAT1 genomic DNA and three sites of T-DNA insertion. Boxes represent exons and lines represent introns. LBb1 and three primer pairs, P1–P2, P3–P4, and P5–P6, were used to identify gfat1-1, gfat1-2, and gfat1-3 alleles, respectively. (B) The frequency of pollen quartets from qrt1-4/qrt1-4 and qrt1-4/qrt1-4 gfat1-2/+ mutant plants bearing zero to four pollen tubes after 16 h incubation. Values represent the means ±standard error (SE) from three independent repeats (n≥100 per repeat). Pollen quartets from qrt1-4/qrt1-4 and qrt1-4/qrt1-4 gfat1-2/+ plants were stained with calcofluor white (C) or with ruthenium red (D). Scale bars: 20 µm (C, D). Pollen grains showing polar deposition of callose and pectin are indicated with arrowheads.