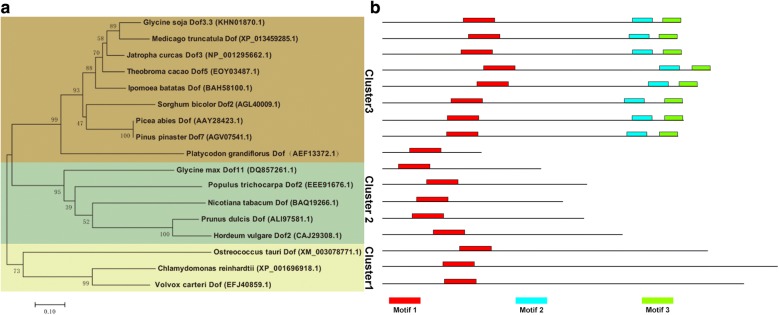
Fig. 2.
a Phylogenetic analysis of crDOF with DOFs from G. max, V. carteri, G. soja, M. truncatula, S. bicolor, J. curcas, P. abies, P. pinaster, I. batatas, P. dulcis, P. grandiflorus, H. vulgare, N. tabacum, T. cacao and P. trichocarpa (sequence IDs listed in Additional file 5: Table S2). The tree was constructed using the neighbour-joining algorithm. Numbers at branch points are bootstrap percentages derived from 1000 replicates. b The MEME motifs are shown in different coloured boxes