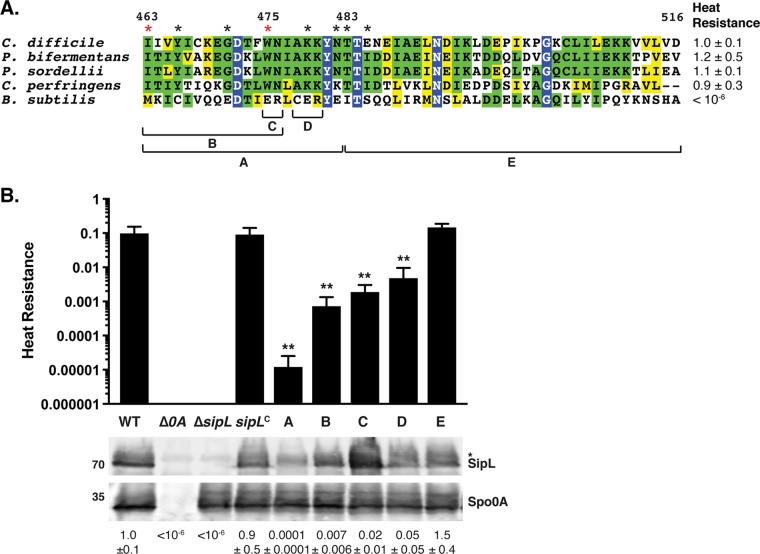
FIG 4.
Chimeric analyses identify a small region of the LysM domain important for SipL function. (A) Alignment of LysM domains from SipL homologs from C. difficile (CAJ70473), P. bifermentans (EQK49575), P. sordellii (EPZ54296), and C. perfringens (ABG83606), as well as the LysM domain from B. subtilis SpoVID (NP_390689). Blue boxes indicate residues that are completely conserved; green boxes indicate residues that are conserved in some of the homologs; and yellow boxes mark residues that are similar between the homologs. The brackets and letters below the alignment indicate the regions swapped between the C. difficile SipL and B. subtilis SpoVID LysM domains. The red asterisks highlight residues whose mutations impaired functional spore formation, while black asterisks indicate residues whose mutation to the residue in the B. subtilis SpoVID LysM domain did not reduce SipL function, i.e., spore formation (Fig. S3). (B) Graphical representation of the heat resistance assay results and Western blot analyses of SipL in the chimeric constructs described for panel A using an antibody raised against SipL lacking the LysM domain. sipLC refers to the ΔsipL/sipL wild-type complementation strain. The asterisk marks a nonspecific band detected by the anti-SipLΔLysM antibody. Spo0A was used as a proxy for measuring sporulation induction (4, 25). The Western blots shown are representative of the results of three independent biological replicates. The heat resistance efficiencies for all chimeric swaps were determined from 20- to 24-h sporulating cultures and represent the mean and standard deviation for a given strain relative to the level of the wild type based on a minimum of three independent biological replicates. Statistical significance relative to results for the wild type was determined using one-way ANOVA and Tukey’s test. **, P < 0.01.