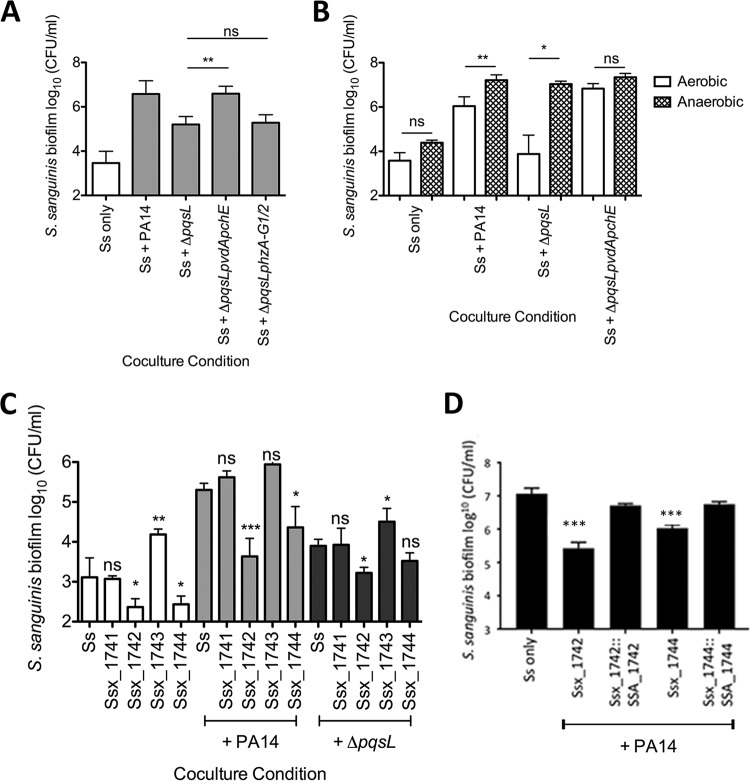
FIG 4.
The P. aeruginosa ΔpqsL mutant likely inhibits Streptococcus growth by sequestering iron via siderophore production. (A) Coculture of S. sanguinis SK36 with P. aeruginosa PA14 mutant strains lacking pqsL and siderophore (ΔpqsL ΔpvdA ΔpchE) or phenazine (ΔpqsL ΔphzA-G1/2) genes. (B) Coculture of S. sanguinis SK36 with P. aeruginosa PA14 mutant strains in 5% CO2 (aerobic) or under anaerobic growth conditions. (C) Coculture of S. sanguinis SK36 gene replacement mutants lacking putative iron acquisition genes with P. aeruginosa PA14 and the ΔpqsL mutant. (D) Complementation assays with the rescued Ssx_1742 and Ssx1744 strains. Each bar represents the average of three biological replicates, each with at least three technical replicates. There was no significant difference between the wild-type S. sanguinis SK36 and the two complemented strains. Error bars indicate SD. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 (by repeated-measures ANOVA with Tukey’s multiple-comparison posttest [A], the paired two-tailed Student t test [B], repeated-measures ANOVA with Dunnett’s multiple-comparisons posttest with S. sanguinis [Ss] as the control condition [C], and the paired two-tailed Student t test comparing each complemented and vector control strain and the paired two-tailed Student t test with a Bonferroni correction with the wild type compared to both complemented strains [D]). The corresponding P. aeruginosa growth data for these experiments can be found in Fig. S11, S13, and S14 in the supplemental material.