Figure 3.
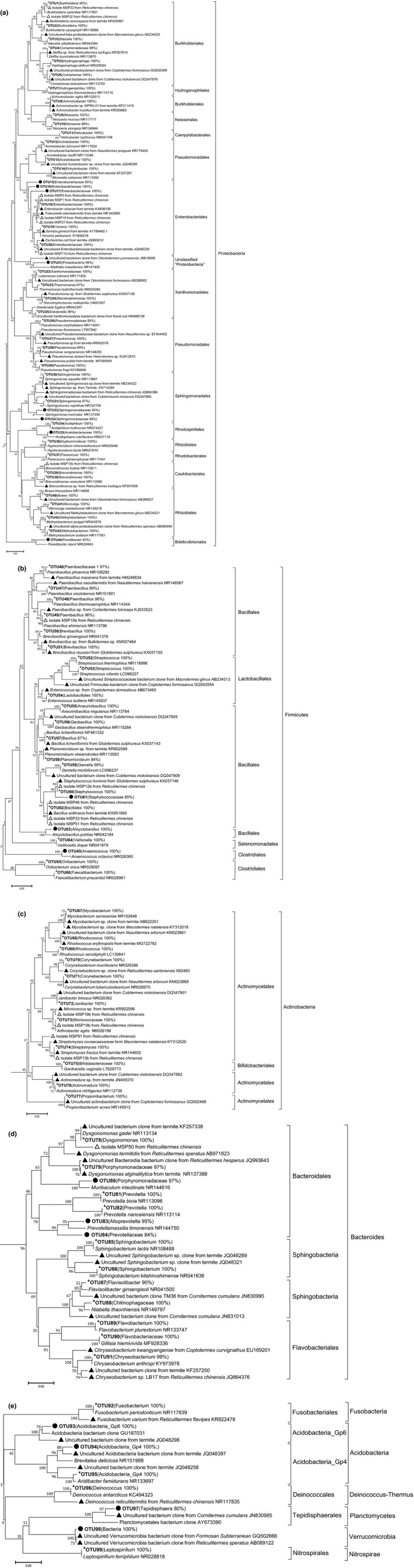
Phylogenetic trees of 99 OTUs from MSP pool. (a) Proteobacteria; (b) Firmicutes; (c) Actinobacteria; (d) Bacteroides; (e) OTUs belonging to other Phyla as indicated. The 16S rRNA sequences of all the published termite‐gut‐derived bacteria were mined from NCBI. The OTU sequences of MSP pool sample were blasted with the GenBank of NCBI and the 16S rRNA sequences of type species with the highest similarity to our OTUs were selected, together with previous mined termite gut derived bacterial 16s rRNA sequences, as reference sequences in phylogenetic tree. Tree viewing was inferred by the Neighbor‐joining method of Mega 6 based on the 16S rRNA gene sequences. OTUs obtained from this MSP pool were showed in bold. The most likely taxon category and its confidence value was listed in the bracket behind each OTU, the confidence threshold was set to be ≥80%. Symbols: ▲: Terminate sequence obtained from GenBank, accession numbers are shown at the end; ●: Sequence with 16S rRNA gene similarity lower than 97% compared with the other isolates in the environments; △: Strains isolated in this study, *: Cultivable taxa.
