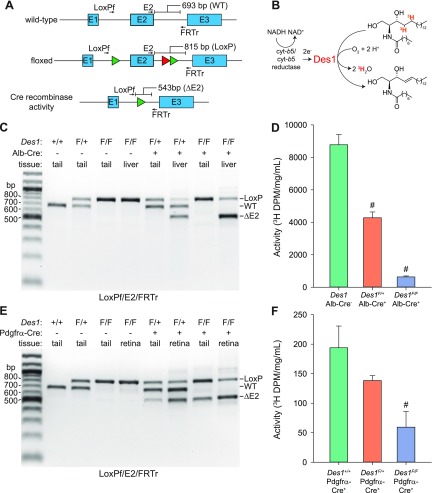
Figure 4.
Genome- and protein-level analysis of efficiency and tissue specificity of Des1 conditional knockout. A) Multiplex PCR strategy for detecting Cre recombinase activity directed toward the floxed Des1 allele. The green and red triangles represent LoxP and FRT sequences, respectively. Primer binding positions are shown as arrows above and below the gene schematics. Primer sequences are given in Supplemental Table S1. B) Assay used to assess the level of Des1 activity in targeted tissues. Des1 activity against a tritiated dihydroceramide substrate releases tritiated water, which can be detected by scintillation counting. C) Multiplex PCR showing the susceptibility of Des1 exon 2 deletion in wild-type or floxed Des1 mice expressing Cre recombinase under the albumin promoter. Primers used for the reactions are listed below the gel image. D) Des1 activity assays conducted by using liver microsome samples from 1- to 2-mo-old mice as the enzyme source. Aqueous radioactivity produced during the reaction was normalized to the total protein concentration in the assay mixture. The “Des1 Alb-Cre” bar shows pooled data from Cre-negative Des1+/+, Des1F/+, and Des1F/F mice, which exhibited comparable activity levels. Des1 Alb-Cre−, n = 3; Des1F/+ Alb-Cre+, n = 3; Des1F/F Alb-Cre+, n = 4. E) Multiplex PCR showing the susceptibility of Des1 exon 2 deletion in wild-type or floxed Des1 mice expressing Cre recombinase under the Pdgfrα promoter. Primers used for the reactions are listed below the gel image. F) Des1 activity assays conducted by using whole retinal lysates, free of RPE contamination, from 1- to 2-mo-old mice of the indicated genotypes as the enzyme source. Aqueous radioactivity produced during the reaction was normalized to the total protein concentration in the assay mixture. Des1+/+ Pdgfrα-Cre+, n = 2; Des1F/+ Pdgfrα-Cre+, n = 10; Des1F/F Pdgfrα-Cre+, n = 4. Data (D, F) are presented as means ± sem. # P < 0.01 vs. Cre negative (D) or wild-type (E) Des1 control groups.