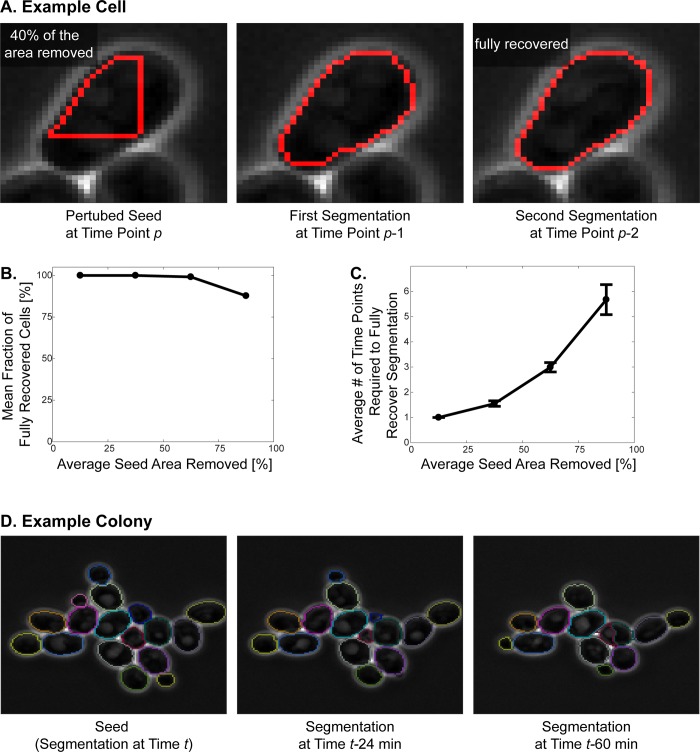
Fig 7. Robustness of the segmentation algorithm.
Robustness to errors in the seed. (A) Example cell: The seed of the example cell is perturbed by randomly removing 40% of the seed. The algorithm uses this perturbed seed to segment the cell at time point p-1 and recovers the cell with only minor mistakes. The algorithm fully recovers the cell in two time points. Note that the algorithm segments the cells backwards in time, thus time points (i.e. frame numbers) are decreasing. (B) The seeds of 340 cells were perturbed by randomly removing 10–90% of the seed. The cells are grouped based on the severity of perturbation, i.e. percent seed area removed, in 25% increments. Mean fraction of fully recovered cells is plotted for each group. Note that out of 340 cells, only 9 of them were not recovered by the algorithm. (C) The cells are grouped based on the perturbation in 25% increments and the average number of time points required to fully recover the correct cell segmentation is plotted for each group. Number of time points required to fully recover the cells increase with the severity of the seed perturbation. The error bars show standard error of the mean. Robustness to time interval between frames. (D) Example colony used for the quantification presented in Table 4. The correct segmentation at time t is used as a seed to segment the images taken at t-24 min and t-60 min. All cells are segmented accurately when the time interval between the seed and the image is 24 minutes. However, when this interval is raised to 60 minutes, a major error is introduced (See the over-segmented cell in red and green.).