Figure 3.
The H1 C-Terminal Tail Stabilizes and Shapes Imp7:Impβ:H1.0
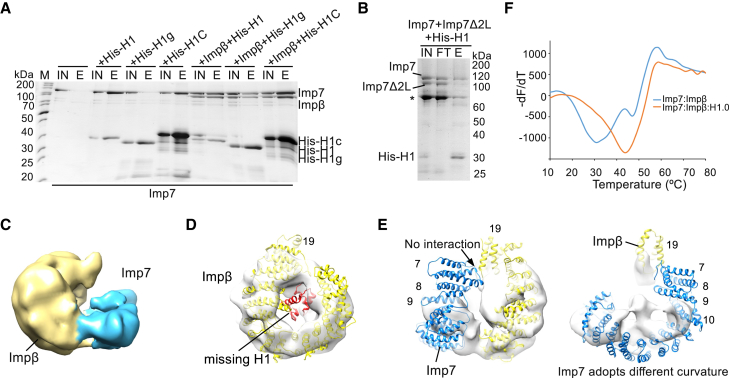
(A) SDS-PAGE analysis of a pull-down experiment. His-tagged wild-type full-length H1.0 or its domains was used as bait to investigate H1 interaction with Imp7 and Impβ. H1g, H1 globular domain; H1C, H1 C-terminal domain; IN, input; E, elution; M, protein marker.
(B) Results of a competition pull-down assay showing that Imp7 acidic loops contribute to H1 binding. His-H1.0 is bound to the nickel-nitrilotriacetic acid (Ni-NTA) resin. Equal amounts of Imp7 and the Imp7Δ2L were added to the assay. The Imp7Δ2L mutant lacks two acidic loops from the unstructured region between HEAT repeats 19 and 20. FT, flowthrough. A degradation product of Imp7Δ2L is labeled with an asterisk.
(C) Cryo-EM map of the Imp7:Impβ complex lacking density for H1.0 resolved to 10.4 Å. Impβ is shown in yellow, and Imp7 is shown in blue. Contour level, ∼0.035.
(D) Model of the Imp7:Impβ:H1.0 complex superimposed onto the Imp7:Impβ map. Impβ and H1.0 are shown in yellow and red, respectively. H1 is not bound by Impβ. Contour level, ∼0.035.
(E) Model of the Imp7:Impβ:H1.0 complex superimposed onto the Imp7:Impβ map. Imp7 and Impβ are shown in blue and yellow, respectively. The N-terminal part of Imp7 is not visible. Imp7 adopts a different curvature. Contour level, ∼0.035.
(F) TSA curves of the Imp7:Impβ:H1.0 and Imp7:Impβ complexes, showing that Imp7:Impβ:H1.0 is more stable than Imp7:Impβ.
See also Figure S5.