Figure 2.
Reactivity of MitoCDNB In Vitro
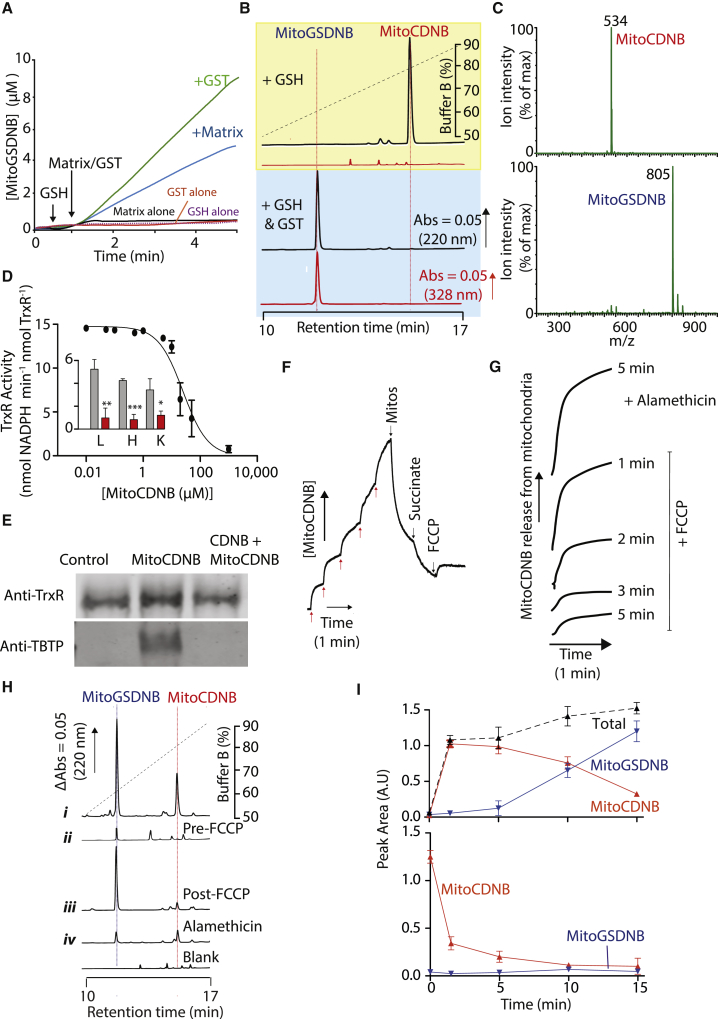
(A and B) GST-catalyzed reaction of MitoCDNB with GSH. (A) MitoCDNB (10 μM) was incubated ± GSH (1 mM) with GST-κ (10 μg) or mitochondrial matrix extract (100 μg protein), and absorbance at 328 nm measured. (B) MitoCDNB (10 μM) was incubated with GSH (1 mM) for 10 min alone (top), or with GST-κ (100 μg, bottom) and then analyzed by RP-HPLC at 220 nm (TPP, blue) and 328 nm (MitoGSDNB, red). Peak identities were confirmed by spiking with authentic compounds (Figure S2D).
(C) Mass spectrometric analysis of MitoCDNB reaction with GSH. MitoCDNB was incubated with GSH (top) or with GSH + GST-κ (bottom) as in (B) above then analyzed by mass spectrometry.
(D) Mammalian TrxR1 and TrxR2 inhibition by MitoCDNB. TrxR1 (25 μg) was incubated with MitoCDNB for 10 min and then assessed for TrxR1 activity. Inset: MitoCDNB inhibition of TrxR2 in matrix extracts (25 μg protein) from rat liver (L), heart (H), or kidney (K) mitochondria, incubated with 5 μM MitoCDNB (red) or vehicle (gray) for 5 min and then assessed for TxR2 activity (units = nmol NADPH min−1 mg protein−1).
(E) Alkylation of TrxR1 by MitoCDNB. TrxR1 (20 μg) was incubated for 10 min with 20 μM MitoCDNB (MitoCDNB), 20 μM CDNB for 5 min followed by 20 μM MitoCDNB for 10 min (CDNB + MitoCDNB) or EtOH control (0.1%). Protein was then assessed by western blotting for TrxR1 (top) and reprobed with anti-TPP antiserum (bottom).
(F) MitoCDNB uptake by mitochondria. An electrode sensitive to the TPP moiety of MitoCDNB was calibrated (5 × 1 μM MitoCDNB, red arrows). Liver mitochondria (2 mg protein/mL) were then added, followed by succinate (10 mM) and 1 μM FCCP. A representative trace is shown of three replicates.
(G) Time dependence of MitoCDNB release from mitochondria upon uncoupling. Mitochondria were incubated with 10 μM MitoCDNB as in (F) and at the indicated times 1 μM FCCP or 5 μg/mL alamethicin was added.
(H) RP-HPLC of mitochondrial MitoCDNB uptake. Liver mitochondria were incubated with 10 μM MitoCDNB as in (F): (i) with MitoCDNB for 9 min; (ii) with FCCP for 4 min followed by MitoCDNB for 5 min; (iii) with MitoCDNB and succinate for 5 min followed by FCCP for 4 min; (iv) with MitoCDNB and succinate for 5 min followed by alamethicin for 4 min. Mitochondria and supernatants (Figure S3C) were then analyzed by RP-HPLC.
(I) Time dependence of uptake and transformation of MitoCDNB. Mitochondria were incubated with MitoCDNB as in (H) and then mitochondrial (top) and supernatant (bottom) fractions analyzed by RP-HPLC for MitoCDNB (red) or MitoGSDNB (blue). Peak areas are in a.u and the normalized sum of the peak areas is in black.
Data are means ± SEM, N = 3. Traces are representative of >3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figures S2 and S3.