Figure 3.
Impact of MitoCDNB on Mitochondrial GSH and TrxR
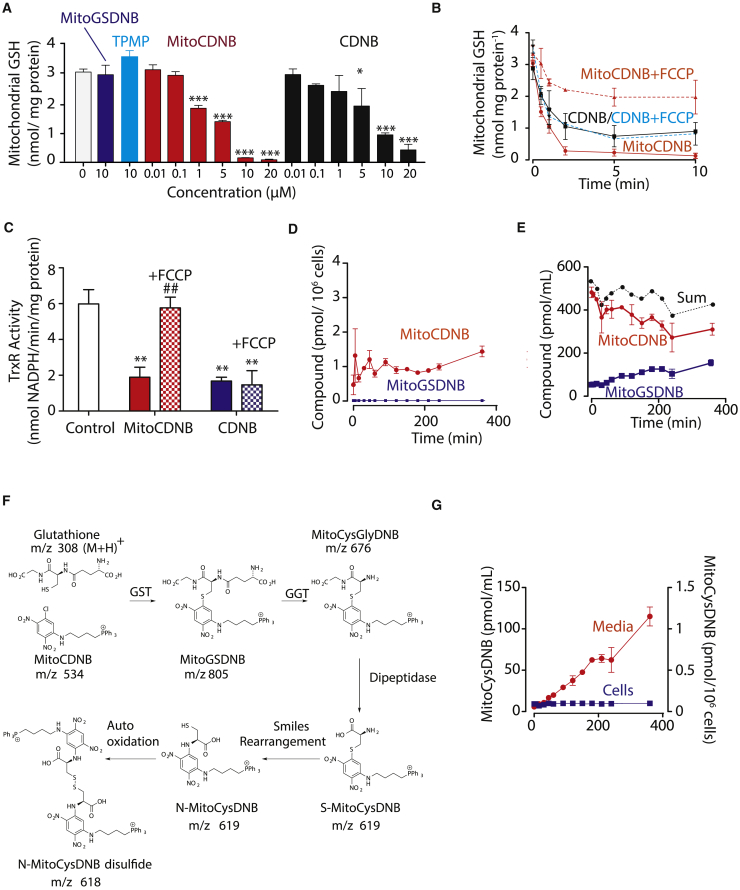
(A) MitoCDNB depletes mitochondrial GSH. Liver mitochondria (2 mg protein/mL) were incubated with vehicle, MitoCDNB, CDNB, TPMP, or MitoGSDNB for 5 min, then matrix glutathione levels were determined.
(B) Time course of GSH depletion. MitoCDNB (10 μM) or CDNB (10 μM) were incubated with mitochondria, then matrix glutathione levels determined. For some experiments FCCP (1 μM) was added prior to MitoCDNB or CDNB (dotted lines).
(C) Inhibition of TrxR2 by MitoCDNB. Liver mitochondria (2 mg/mL) were incubated for 5 min with either carrier (white), MitoCDNB (10 μM, red), or CDNB (10 μM, blue). For some experiments FCCP (1 μM) was added before MitoCDNB (red, white) or CDNB (blue, white) for a further 5 min.
(D and E) Uptake and metabolism of MitoCDNB by cells. HepG2 cells were incubated with MitoCDNB (10 μM) and then the MitoCDNB and MitoGSDNB in the cells (D) and medium (E) were quantified by LC-MS/MS. The sum of MitoCDNB and MitoGSDNB is indicated.
(F) Schematic of MitoCDNB metabolism. MitoCDNB reacts with GSH to form MitoGSDNB in mitochondria. This is excreted and broken down by the extracellular γ-glutamyl transpeptidase (GGT) to MitoCysGlyDNB, further converted to S-MitoCysDNB, which undergoes a spontaneous Smiles rearrangement to N-MitoCysDNB, which autooxidizes to N-MitoCysDNB disulfide.
(G) Generation of MitoCysDNB in HepG2 cells. MitoCysDNB levels were quantified in cells from (D) above and from media (E). Data are means ± SEM, N = 3.
*p < 0.05, **p < 0.01, ***p < 0.001 relative to control; ##p < 0.01 relative to incubation without FCCP in (C) only. See also Figures S3 and S4.