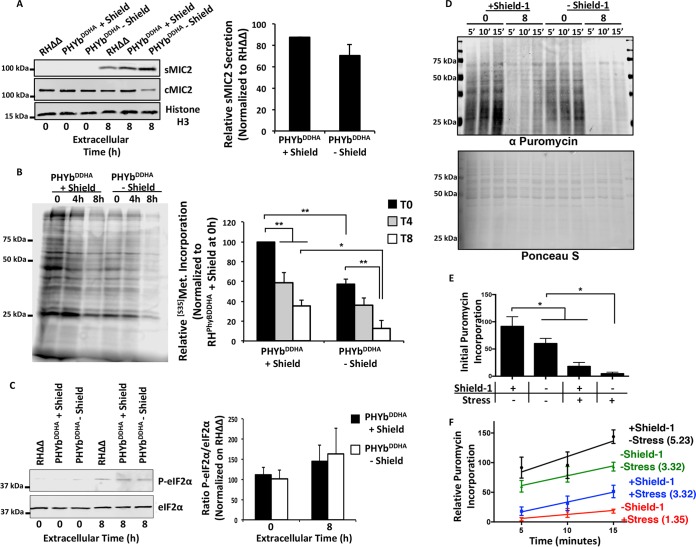
FIG 6.
TgPHYb is required for de novo protein synthesis during extracellular stress. TgPHYbDDHA parasites grown for 24 h with or without Shield-1 were harvested and exposed to stress for 0, 4, or 8 h at 21% O2. (A) Parasites were pelleted by centrifugation. MIC2 was detected in equivalent volumes of the pelleted parasites (cMIC2) and supernatants (sMIC2) collected from the parasites following 8 h of incubation. Toxoplasma histone H3 was used as a loading control. Shown are the average and standard deviations from 3 independent experiments as well as representative blots. (B) Autoradiograph of parasites pulse-labeled with [35S]Met-Cys for 1 h following exposure to extracellular stress for 0, 3, or 7 h. Shown are the average and standard deviations from 3 independent experiments as well as representative blots. *, P < 0.05; **, P < 0.005, one-way ANOVA. (C) Western blot detection of phospho-eIF2α or total eIF2α in parasites exposed to extracellular stress for 0 or 8 h. Shown are the average and standard deviations from 3 independent experiments as well as representative blots. A Student t test indicated that differences were not statistically significant. (D) Freshly egressed parasites or parasites incubated extracellularly for 8 h were incubated with puromycin for the indicated times. Lysates were prepared, separated by SDS-PAGE, and then either Western blotted to detect puromycylated peptides (top) or stained with Ponceau S (bottom). Shown are representative blots from 3 independent experiments. (E) Initial puromycin incorporation levels were determined by ratios of puromycin to Ponceau S staining intensities after 5 min of adding puromycin. Data were normalized to unstressed parasites in the presence of Shield-1. Shown are averages and standard deviations from 3 independent experiments. *, P < 0.05, Student’s t test. (F) Puromycin/Ponceau S labeling intensities were determined for each time point, and the slope (shown as number on label) of each line was calculated to determine rate of puromycin incorporation.