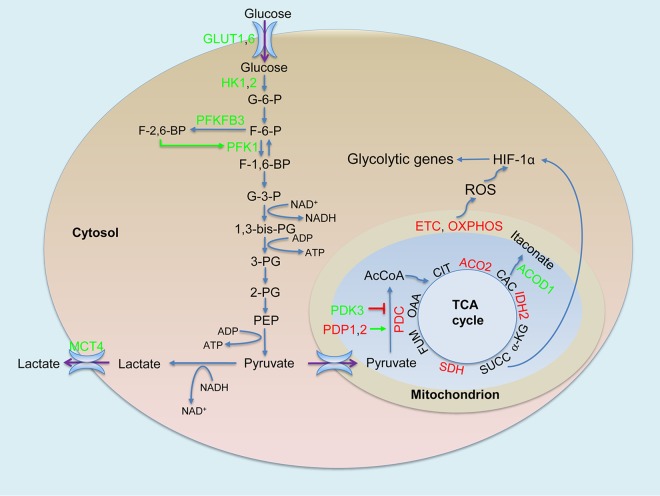
FIG 1.
The Warburg effect in M. tuberculosis-infected macrophages. The early phase of macrophage infection is accompanied by increased glycolytic flux with consequent lactate formation and secretion and, concurrently, decreased mitochondrial oxidative metabolism exemplified by decreased pyruvate oxidation by the pyruvate dehydrogenase complex (PDC) and the downregulation of the TCA cycle and oxidative phosphorylation (OXPHOX). Increased levels of PFKFB3, one of the key regulatory mechanisms to promote glycolytic flux, together with elevated levels of glucose uptake transporters (GLUT1 and GLUT6) and other glycolytic enzymes, including hexokinases (HK1 and HK2), type 1 phosphofructokiase (PFK-1), and lactate secretion transporter member 4 (MCT4), illustrate the occurrence of the Warburg effect. The Warburg effect is associated with increased expression and activity of HIF-1α caused by the production of reactive oxygen species (ROS) derived from the respiratory electron transport chain (ETC) and OXPHOX and by the succinate (SUCC) accumulation from the TCA cycle in mitochondria. Green, increased expression/activity; red, decreased expression/activity; green arrow, stimulation, red block line, inhibition. Abbreviations: PDK3, pyruvate dehydrogenase kinase 3; PDP1-2, pyruvate dehydrogenase phosphatase 1 and 2; PDC, pyruvate dehydrogenase complex; ACO2, aconitase 2; ACOD1, aconitate decarboxylase 1; IDH2, isocitrate dehydrogenase 2; SDH, succinate dehydrogenase; G-6-P, glucose-6-phosphate; F-6-P, fructose-6-phosphate; G-3-P, glyceraldehyde-3-phosphate; 1,3-bis-PG, 1,3-bis-phosphoglycerate; 3-PG, 3-phosphoglyceride; 2-PG, 2-phosphoglyceride; PEP, phosphoenolpyruvate; AcCoA, acetyl CoA; OAA, oxaloacetate; CIT, citrate; CAC, cis-aconitate; α-KG, α-keto-glutarate; FUM, fumarate. Web-based GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used for differential gene expression analysis, and changes described correspond to an adjusted P value of <0.05 at corresponding time points relative to uninfected controls. Microarray data are from the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo/; accession numbers GSE79733 and GSE31734).