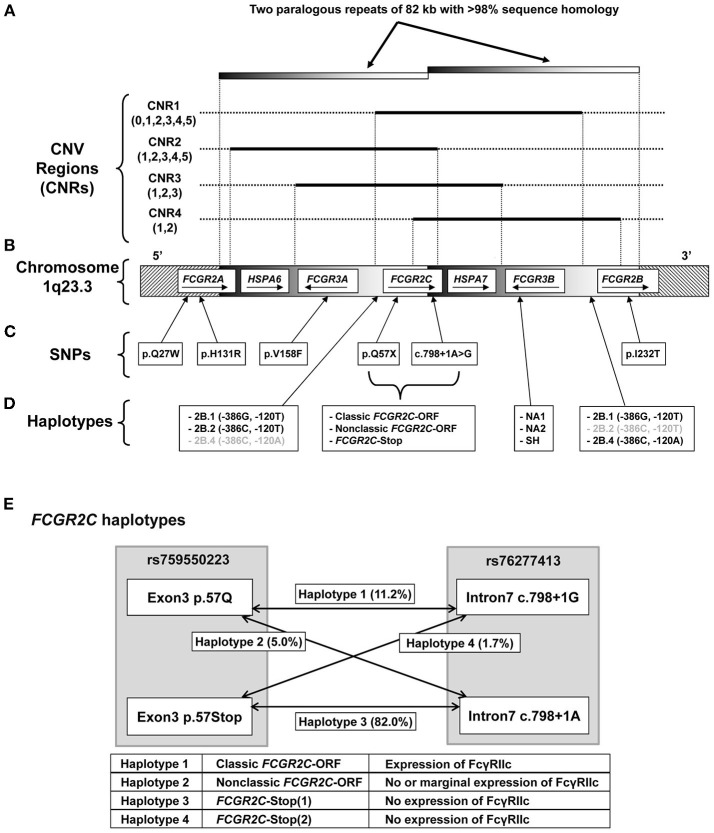
Figure 1.
Genomic organization of the FCGR2/3 locus. Overview of the FCGR2/3 locus at 1q23.3. (A) CNV at the locus occurs always in regions containing a series of genes, termed CNV regions (CNRs) (18, 21). Numbers between brackets indicate observed copy number for that CNR in the individuals tested in this study, black lines indicate the extent of the different CNRs. Gray shaded bars indicate the extent of two paralogous repeats of the locus. The novel rare CNR4 that we recently described (18) was found in 4 of the individuals included in this study, and was combined with the similar CNR1 in this study for reasons of simplicity. (B) Overview of the genes in the locus with their orientation. Depiction of genes is not to scale. (C) Functional SNPs at the locus, indicated by single letter amino acid code and amino acid position, except for the splice variant c.798+1A>G. All these SNPs were determined in this study. (D) Functional haplotypes at the locus. 2B.1, 2B.2, and 2B.4 are haplotypes of two SNPs (at nucleotide positions −386 and −120 relative to the start of translation) in the otherwise identical promoter regions of FCGR2B and FCGR2C (haplotypes in gray are very rarely found in that particular gene). FCGR3B NA1, NA2, and SH are haplotypes determined by six SNPs. These haplotypes determine the different allotypes of Human Neutrophil Antigen 1 (HNA1) involved in neutrophil alloimmunization, and respectively encode the HNA1a/HNA1b/HNA1c antigenic variants. (E) Schematic representation of the different haplotypes of FCGR2C haplotypes, which determine expression of FcγRIIc. FCGR2C-Stop (1) and FCGR2C-Stop (2) haplotypes are similar in function and expression and are taken together as FCGR2C-Stop throughout the manuscript. Percentages represent allele frequencies of the different haplotypes in European healthy controls.