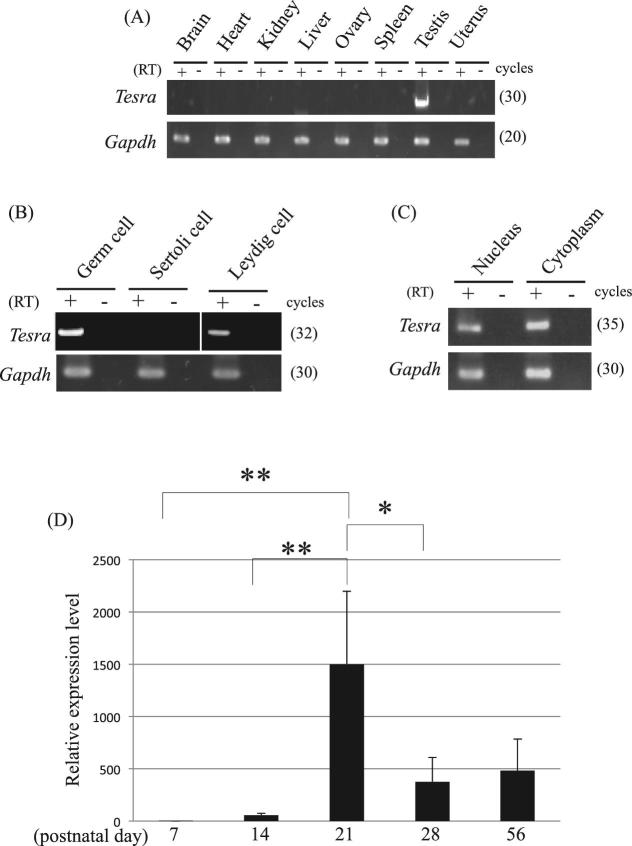
Figure 2.
Expression pattern of Tesra. (A) Tissue specificity of Tesra. Eight mouse tissues, as indicated, were collected from adult mice and used for RT-PCR. Reverse transcription was performed by using the oligo(dT) primer with (RT+) or without reverse transcriptase (RT–). Gapdh was detected as an internal control. The cycle number of each reaction is indicated in parenthesis. (B) Cell type specificity of Tesra in the testis. Mouse germ, Sertoli, and Leydig cell fractions were prepared and used for RT-PCR. Germ cells and Leydig cells were collected from adult mouse testes, while Sertoli cells were collected from 7- to 12-day-old testes and cultured for several days. The experiment was done and the data are presented as in (A). (C) Subcellular localization of Tesra in mouse male germ cells. Germ cells were collected and fractionated into nuclear and cytoplasmic fractions, and total RNAs were purified from both fractions. Gapdh was a positive control, in which both forward and reverse primers were designed within exon 6 of the gene. RT-PCR was done and the data are presented as in (A). The Tesra transcript was detected in both nuclear and cytoplasmic fractions. (D) Tesra expression during postnatal testis development. Mouse testes at postnatal days 7, 14, 21, 28, and 56 were collected, and total RNAs were purified and used for qRT-PCR analysis. The housekeeping Aip gene was examined as an internal control. The level of Tesra was normalized to Aip, and the value at day 7 was set to 1.0. The data are presented as means ± SD from three independent experiments, and the statistical significance was analyzed by one-way ANOVA followed by the Tukey post hoc test. *P < 0.05. **P < 0.01.