Scheme 1.
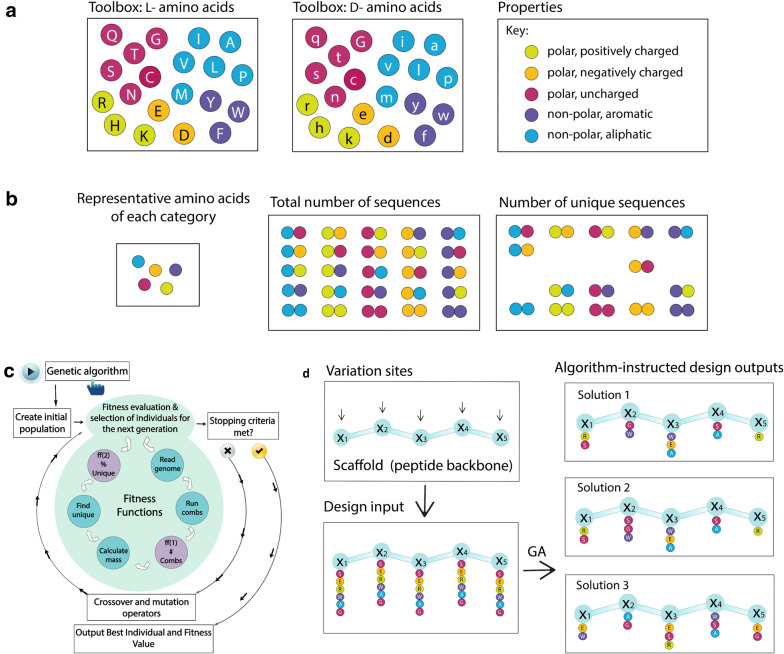
Towards the algorithm-supported design of random peptide libraries. a Amino acid toolbox of L- and D-stereoisomers of 20 proteinogenic amino acids color-coded to represent a specific property: hydrophobic-aromatic (magenta), hydrophobic-aliphatic (blue), hydrophilic-uncharged (purple), hydrophilic-positively charged (green), and hydrophilic-negatively charged (orange). b Graphical representation of building an OBOC peptide library, based on dipeptides (r = 2), using the representative amino acids from the toolbox (m = 5), depicting the advantage of working with sequences having unique masses. The calculator explores the full sequence space () and excludes permutations that present the same mass and differ only in the amino acid order within the sequence. c Schematic representation of the 2-objective genetic algorithm used to perform the optimization, indicating the steps required to perform the selection of individuals for the next generation. The main criteria of the GA are two fitness functions for maximizing (1) the number of all amino acid permutations and (2) the number of sequences having unique mass. d Schematic representation of the optimization algorithm input and outputs suggesting the best design options (for each position r) to obtain maximally diverse random peptide libraries