Figure 3.
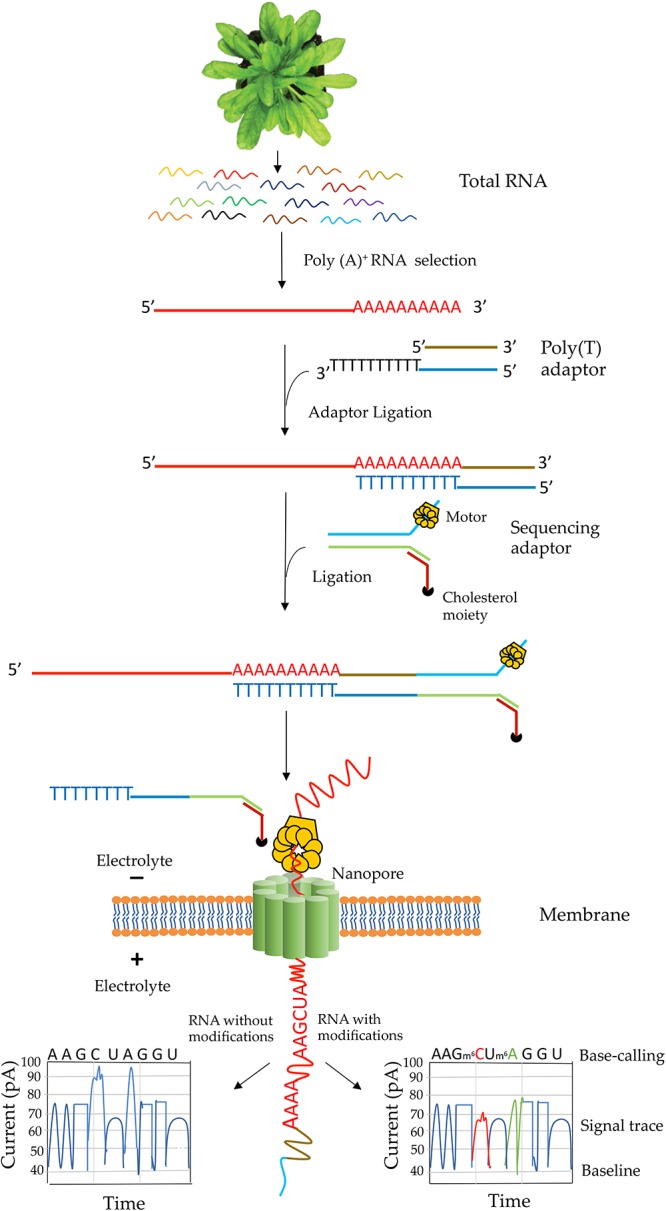
Schematic illustration of direct RNA sequencing using the Oxford Nanopore Technology. Poly(A)+ mRNA from total RNA is isolated, then a poly(T) adaptor and a sequencing adaptor with a motor enzyme are added to the 3′ end of poly(A)+ mRNA. It is then subject to sequencing on a membrane with thousands of nanopore channels, each of which is coupled to ammeters that measure current passing through the pore. The motor enzyme interacts with a nanopore on an electrically resistant synthetic membrane and the RNA strand is fed through the nanopore. A voltage across the membrane is applied and as the RNA moves through the nanopore nucleotide bases cause a characteristic change in current through the pore that is unique to each normal and modified base. The current output is then used in base-calling. An example of current output when RNA with (right box) or without modified RNA bases (left box) move through a pore is shown.
