Abstract
An emerging concept in cell signaling is the natural role of reactive oxygen species, such as hydrogen peroxide (H2O2), as beneficial messengers in redox signaling pathways. Despite growing evidence, the nature of H2O2 signaling is confounded by difficulties in tracking it in living systems both spatially and temporally at low concentrations. Here we develop an array of fluorescent single-walled carbon nanotubes that can selectively record in real time the discrete, stochastic quenching events that occur as H2O2 molecules are emitted from individual human epidermal carcinoma cells that are stimulated by epidermal growth factor (EGF). We show mathematically that such arrays can distinguish between molecules originating locally on the cell membrane from other contributions. We find that EGF induces 2 nmol H2O2 locally over a period of 50 min. This platform promises a new approach to understanding the signaling of reactive oxygen species at the cellular level.
Historically, H2O2 is thought to have only a deleterious role in cell biology as a toxic metabolic waste product, or as part of the immune respiratory burst in response to microbial invasion1. New findings suggest that it is a messenger in normal signaling pathways: H2O2 is produced when cells are stimulated with various growth factors, cytokines and other signaling molecules, and is known to activate specific downstream targets2. Understanding the role of H2O2 and other reactive oxygen species (ROS) is hampered by their low concentration and short lifetime. This has inspired innovative detection probes3–7 but many still cannot map the entire transient signaling response on a single molecule level over its duration with spatial resolution6–8.
Our laboratory has pioneered the use of single-walled carbon nanotubes (SWNT) as fluorescent optical sensors for analytical detection from within living cells and tissues. Recent measurements by others9 and our own laboratory10–11 have extended the detection limit down to the single-molecule level by analyzing the stochastic quenching of excitons as molecules adsorb to the SWNT surface. Here we develop an array of such single-molecule sensors selective for H2O2 that can image the signaling flux emanating from single living A431 human epidermal carcinoma cells in real time with spatial precision. These arrays can resolve several questions about H2O2 generation upon growth factor stimulation, including the membrane activity compared to the other contributions, its spatial distribution, and through inhibition experiments, the chemical mechanism of the signal response.
A431 cells over-express the epidermal growth factor receptor (EGFR), which is one of four transmembrane growth factor receptor proteins12. EGFR is a 170-kDa glycoprotein with an extracellular receptor domain, a transmembrane domain and an intracellular domain13. The extracellular domain is divided into four subdomains: I, II, III and IV, with I and III participating in binding14 (Fig. 1a). A431 cells express15 approximately 106 EGFRs per cell while only 4×104 to 105 receptors occur per non-cancerous cell16. Epidermal growth factor16 (EGF) - a single polypeptide chain of 53 amino acid residues held together by three disulfide bonds in cysteine - stimulates cell growth, proliferation and differentiation upon binding to EGFR (Fig. 1a)16. Upon activation by EGR, EGFR undergoes dimerization at the cell membrane13 and an H2O2 signal is generated17. The chemical origin of this H2O2 signal and its relationship to the remaining cascade is largely unknown.
Figure 1.
Nanotube sensing platform. a, A431 cell cultured on collagen-SWNT film. Zoom in on red circle: EGFR domains spanning the cell membrane. Domains I and III bind to EGF (red) and generate H2O2. b, NIR image of SWNT underneath A431 (left) and phase contrast image of A431 cell (right) cultured on SWNT sensors (658 nm excitation, 1mW, Alpha Plan-Apo 100x/1.46 oil emersion objective). c, Forward and reverse binding rate of SWNT sensor for various analytes show selectivity for H2O2. d, Fluorescence trace for control (no cells) show no steps. e, Trace for SWNT in the red circle in (b) show reversible, stepwise quenching (green trace), modeled by HMM (red).
Recent progress in fluorescent probes has confirmed the generation of H2O2 signal in A431 in response to EGF, and has also identified a similar mechanism in neuronal cell signaling6–8. Our work differs in its single-molecule detection limit, and infinite photoemission lifetime allows continuous detection over the entire response in real time. The array of single-molecule sensors can mathematically discriminate signals generated at the membrane (near-field) from those originating from the cell interior (far field), an important property for cell analysis. Mapping shown before and after EGF stimulation of A431 cells and NIH-3T3 murine fibroblast cells in real time, informs the chemical mechanism of the signaling cascade.
Selective SWNT sensors detect and map H2O2 efflux spatially
The SWNT array is ideally suited for imaging H2O2 fluxes from living cells because H2O2 binds with a forward (77.8 M−1s−1) and reverse (0.0006 s−1) rate constant10 that allows sensitive detection and that excludes other ROS with long lifetimes. Previously10, we showed that Hidden Markov Modeling can determine forward and reverse rate constants from single-molecule adsorbates on SWNT. Figure 1c is the application of the technique to several species of interest in this work. Except for nitric oxide (NO), H2O2 has the largest forward rate constant of all, meaning its capture probability is the highest. Protons (H+) can be detected with a forward rate of 8.1 M−1s−1, but their reverse rate constant is high (0.0011 s−1) such that, at physiological pH (7.4) their contribution is negligible. Likewise, interfering nitrites and nitrates have small rate constants and there is no contribution from components of cell media. NO has a high forward binding rate of 80.0 M−1s−1, however its reverse rate is almost immeasurable under these experimental conditions. This means that its presence can be easily distinguished, as it irreversibly deactivates the SWNT single-molecule sensor that it encounters. Excluding such signals can subtract the contribution of NO, however no such events were measured in this work, indicating NO was not detected. In addition, our sensor is inert to singlet oxygen (1O2) and superoxide () (Fig. 1c).
Our single-molecule detector array is embedded in a thin film, with 2 nm roughness (Fig. S1) and open porosity (average pore size = 30 nm) towards the SWNT. Therefore, only the most stable species emanating from the cell are detected. For example, 1O2, and OH· have lifetimes of 4 μs18, 1 μs and 1 ns respectively19. These species are not likely to diffuse into the film and interfere with SWNT. The selectivity of our film, together with the diffusion calculation, result in an array of sensors specifically designed to detect single H2O2 molecules in real time. We note that this does not limit the approach exclusively to H2O2. We recently showed that by varying the chemistry of the encapsulating matrix20, or by utilizing multiple orthogonal optical responses (multi-modality)11, it is possible to selectively detect virtually any single-molecule analyte emanating from the cell. Future work will underscore this point.
The detection limit for this class of single-molecule sensor is exchanged for observation time (Fig. S2). For instance, an array of SWNT can detect a concentration of 1 μM (10 μM) provided an acquisition time of 14 min (3 min).
When no cells are plated onto the collagen-SWNT array, the result is a photoluminescence intensity of constant root mean square value. A Hidden Markov algorithm finds no quenched states outside of the noise floor as expected. SWNT sensors near or underneath plated A431 cells show discrete quenching transitions of the type observed previously9–10. Stepwise quenching and dequenching reactions are clearly observable (Fig. 1e), compared to the control experiment (Fig. 1d). Because the collagen-SWNT array has such high selectivity towards H2O2, we assign this flux as single H2O2 originating from cellular metabolic activity2 and also non-specific receptor-ligand binding21. To further support this assignment, we use manganese oxide (MnO2) to selectively catalyze the decomposition of H2O2 around the A431 cells both with and without EGF stimulation, to show that the quenching reverses significantly (Fig. S3) as H2O2 is depleted.
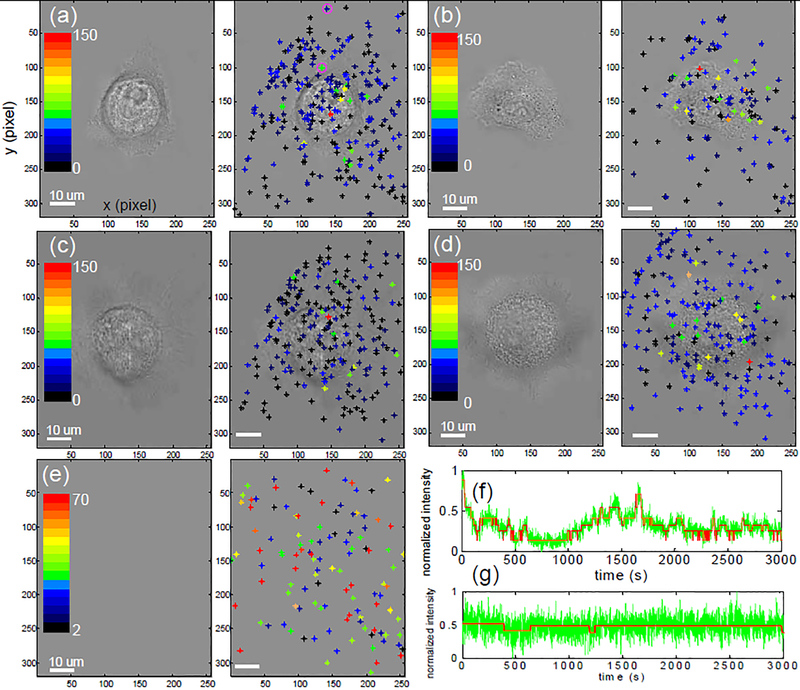
Hidden Markov algorithm was applied to each SWNT signal in the array, yielding the spatial and temporal detection of single molecules emitted from the cell in real time. The typical observation time was 3000s and Figure 2a–d describes the spatial distribution of detection frequencies for both live (Fig. 2a, b) and fixed (Fig. 2c,d) A431 cells after the addition of EGF (500ng/mL) at t = 0 using a Matlab program written by us. Each sensor was binned according to its number of quenching transitions within the 3000s observation window into one of 16 color categories between 0 and 150 counts for Fig 2a–d and between 2 and 70 counts for Fig 2e. Note that the control array, exposed to 10 μM H2O2 in the absence of cells, demonstrates a spatially random distribution of transition frequencies (Fig. 2e). However, when instead A431 cells are present, the frequency distribution possesses a sharp mode invariably confined to the region immediately under the cell. The behavior is seen for both live and fixed cells. The locations of these “hot spots” do not remain invariant over the course of the 3000 s experiment, but shift to alternate regions. The EGFR lifetime15 is approximately 30 min, long enough to prevent spatial averaging of the membrane signal. Example fluorescence traces from a nanotube right under the cell (green star, highlighted in pink circle, Fig. 2a) and nanotube far away from the cell (dark blue star, highlighted in pink circle, Fig. 2a) is shown in Fig. 2f and g. It is clear from these examples that the dominant contribution of the H2O2 flux comes from the A431 cells, and the data suggests that at any given time it is concentrated at specific locations on the array, unlike the case of a uniformly exposed control.
Figure 2.
Spatial mapping of quenching transitions over single A431 cells. a-d, Quenching activity (unit of counts) over the 3000s observation window of each sensor was binned into sixteen categories represented by 16 different color bars with red having the highest quenching activity and black the lowest for live (a, b) and fixed (c, d) A431 cells. e, Control experiment where 10μM H2O2 was present in the absence of a cell. Left panels show phase contrast images without the overlap of quenching activities. Fluorescence trace of green star (f) and dark blue star (g) from (a) are shown.
Real-time quantitative analysis of EGF stimulation
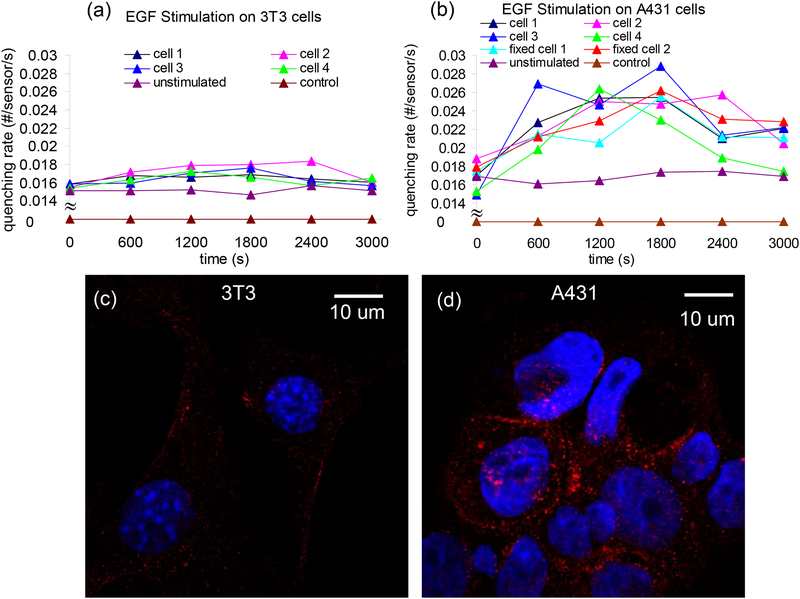
First, we analyze the total dynamic count rate of each single cell in response to EGF to measure the duration of the induced efflux. The quenching rate was calculated in real time for EGF stimulation (500ng/mL EGF was added at t=0) on live and fixed 3T3 (Fig. 3a) and A431 cells (Fig. 3b). Compared to the no cell control and unstimulated cell data, the quenching rate of both A431 and 3T3 cells were increased by EGF stimulation. A431 cells with a higher EGFR density have a much higher quenching rate than 3T3 cells. As can be seen from Fig. 3a–b, the behaviors of single A431 cells after EGF stimulation are similar: the quenching rate increased rapidly right after stimulation. However, the time point of maximal response ranges from 600s to 1800s after stimulation. There is no significant difference between live and fixed A431 cells. Removal of EGF decreases the quenching (Fig. S4). Compared to ensemble measurements on thousands of cells, our platform allows real-time quantification on single isolated cells for the first time. To confirm that the above results correlated with overexpression of EGFR, we then compared the EGFR density in 3T3 cells and A431 cells via immunostaining (Methods). As can been seen in the confocal images (Fig. 3c, d), A431 cells express much more EGFR than 3T3 cells (see Fig. S5 for more immunostaining images). From a calculation on the immunostaining images of 100 cells, the EGFR density of A431 cells is approximately 10 times that of 3T3 cells, consistent with the literature15–16.
Figure 3.
SWNT quenching depends of EGFR density. a, b, Real time quenching rate for live 3T3 cells (a) and live/fixed A431 cells (b). The number of sensors under each single cell is 255, 200, 250, 150, 255, 200 respectively for cell 1, 2, 3, 4, unstimulated and control in (a); 160, 110, 126, 174, 140, 180, 180, 200 respectively for cell 1, 2, 3, 4, fixed cell 1, 2, unstimulated and control in (b). Representative confocal images for 3T3 cells (c) and A431 cells (d) with EGFR (red) labeled with rabbit polyclonal antibody against EGFR and Alexa Fluor 568 donkey anti-rabbit IgG. Nuclei (blue) is stained with 4’,6-diamidino-2-phenylindole (DAPI).
Near-field generation from the membrane
Rank ordering the sensor responses from lowest to highest capture rate constructs the cumulative distribution (supplement). Let x be the number of sensors having a response less than y, so that is then the probability of finding a sensor with a number of counts less than y. For the case of equal capture probabilities, the rank-ordered response is a modified Gamma distribution (Fig. S6a–b):
| (Eq. 1) |
Where ϒ, Γ and P represents the lower incomplete, ordinary and regularized Gamma functions respectively with a as the mean value of y.
A kinetic Monte Carlo simulation of 104 H2O2 molecules randomly binned into a sensor array consists of 300 sensors (n=300) is well-described by Eq. 1 after rank ordering (Fig. S6a–b). Note that all simulations (no fit parameters) utilized Matlab and were repeated 100 times with the average reported unless specified otherwise.
An array of sensors capable of detecting discrete, single molecules has the following unique property: it is possible to distinguish between those near-field component generated at the interface and those comprising a far-field component with no memory of origination.
On top of the far-field component described by Eq. 1, molecules generated near the array surface (i.e. at the cell membrane surface) are easily distinguished. The algorithm for extracting this interfacial generation at the interface simply accounts for non-binomial contributions to the frequency distribution. The local response, ylocal(x), is:
| (Eq. 2) |
The mean value a can be found by computing the slope of the experimental data in the x→ 0 limit:
It can be shown from Monte Carlo simulation that membrane generation near the array interface of sufficient activity always biases the rank-ordered response to the highest activity sensors (Fig. 4a–b, Fig. S6e–i). As a result, a small number of data points at x→ 0 are enough to extract the far-field component (Eq. 1) from any experimental curve with the membrane generation recovered from Eq. 2. A practical sensor array constructed as described above has a distribution of sensor capture probabilities as each sensor varies slightly in length and orientation. A beta distribution is a generic, empirical function that can describe this variation. It has the advantage that the far-field component then becomes a cumulative beta binomial distribution (see simulation and fit in Fig. S6c–d), and an analogous deconvolution can be derived (Fig. 4c–d). The rank-ordered sensor responses of SWNT-collagen arrays exposed to constant (uniform) concentrations of H2O2 from 10 to 100 μM are described by beta binomial distributions with parameters α =1.2 ± 0.15 and β = 3.0 ± 0.12 (Fig. S7a–d). These parameters were used to correct the measured responses for the variation of capture sensitivities of each SWNT.
Figure 4.
Quantitative analysis of results from SWNT sensor array. a-d, Simulation of sensor response (a), rank-ordered sensor response from a (b), sensor response following beta distribution (c) and rank-ordered sensor response from c (d) of 105 H2O2 randomly falling onto 300 sensors (blue), with additional response to local generation (red). After far-field component subtraction from the rank-ordered sensor response (black, -EGF; green, +EGF), the local generation before (blue, star) and after (red, star) EGF stimulation for live (e), fixed (f) A431 cell and live 3T3 cell (g). h, Real-time quenching rate for fixed A431, before (green) and after (blue) EGF stimulation. Sodium azide decreases the quenching, with (red) and without (black) EGF. Extending the singlet oxygen lifetime using D2O increases the quenching (purple). Concentration profiles on log-log scale for different species from solving Reaction 1–8 (i) and from considering the effect of NaN3 when solving the reaction network (j). k, Scheme of the proposed pathway for H2O2 generation.
Membrane activities on single live, fixed A431 cells and live 3T3 cells before and after EGF stimulation over 3000s were extracted from the above algorithm (Fig. 4e–g, see Fig. S8 for the whole data set). Before stimulation, the local activity is negligible. After stimulation, the membrane generation observed for both live and fixed A431 cells, however not for live 3T3 cells. The increased activities after stimulation in the unit of number of quenching transitions per sensor are summarized in Table 1. For A431 cells after simulation, the local H2O2 concentration is determined through calibration to be 2μM using control experiments in the cell-free system (Fig. S7e). The local generation rate from each membrane source is then 0.04 nmol H2O2/min after correcting for diffusion from Eq. S3.
Table 1.
Number of quenching transitions per sensor from receptors alone calculated for live, fixed A431 cells and live 3T3 cells.
| Cell type | Number of quenching transitions per sensor |
|---|---|
| Live A431 cell #1 | 6.6 |
| Live A431 cell #2 | 4.0 |
| Live A431 cell #3 | 5.0 |
| Live A431 cell #4 | 7.5 |
| Fixed A431 cell #1 | 4.5 |
| Fixed A431 cell #2 | 7.8 |
| A431 cells average | 6.0 |
| Live 3T3 cell #1 | 1.0 |
| Live 3T3 cell #2 | 1.1 |
| Live 3T3 cell #3 | 1.1 |
| Live 3T3 cell #4 | 1.0 |
| 3T3 cells average | 1.1 |
A consistent H2O2 signal generation mechanism
The spontaneous or catalytic breakdown of is considered to be the source of H2O2 in many biological pathways, not only for immune cells, but also in a variety of eukaryotic cells2. can be produced by the partial reduction of oxygen by cytochrome c oxidase in mitochondria2 or by membrane-associated NAD(P)H oxidase6. Extensive literature has shown that EGF stimulated H2O2 generation originates from NAD(P)H oxidase instead of mitochondria for various nonphagocytes, including A431 cells6, 22–23. Growth factors like EGF induce the formation of a complex on NAD(P)H oxidase to promote the electron transfer from NAD(P)H to molecular oxygen23. Fixation of A431 cells using 4% paraformaldehyde were designed to remove the influence of the mitochondria24 in our experiments, as has been used in the literature to kill the cells without affecting the binding abilities of EGFR21. The fact that we see no quantitative difference between live and fixed A431 cells in their EGF inducible membrane generation (Table 1) is consistent with the all existing literature where mitochondria do not affect the EGF-induced H2O2 generation. In addition, our single-molecule sensor array allows the membrane signaling flux to be differentiated from a diffuse far-field component for the first time, and can therefore inform the discussion of the nature of the H2O2 signal. Our analysis above confirms the H2O2 that increases in response to EGF binding is generated at the membrane and not in the cell interior (Fig. 4e–g). Recently, DeYulia and co-workers demonstrated that the H2O2 production is EGFR-ligand-dependent in A431 cells21, where the inhibition of EGFR phosphorylation did not affect the H2O2 generation. At this point, both the signaling network post-NAD(P)H oxidation, and the connection between activation of NAD(P)H oxidase and EGFR-ligand-dependent generation, are unclear from the literature. We further performed inhibition experiment using NAD(P)H oxidase inhibitors(Fig. S9a–b) and EGFR inhibitor (Fig. S9c) and found that consistent with our speculation, NAD(P)H oxidase inhibitors prevent H2O2 from forming while EGFR inhibitor has no effect on the H2O2 produced.
It is also not clear what catalytic portion of EGFR may be responsible for membrane H2O2 generation from previous work21. Tryptophan (Trp) is proposed to be responsible for the conversion of 1O2 to H2O2 in antibodies25. This antibody-mediated process is triggered upon binding of 1O2 to conserved binding sites within the antibody fold26, where the antibody serves as the catalyst, stabilizing the intermediate (H2O3) and directing its conversion to H2O2. Trp is present in both EGF16 (Trp 49, Trp 50) and EGFR27 (Trp 140, 176, 453, 492) (Fig. 1a). If lacking these Trp residues, EGFR does not bind ligand with high affinity27. It is possible that EGFR, upon binding with EGF, allows greater access to sites on the receptor itself that catalyze the conversion of 1O2 to H2O2. To explore this, 1 mM sodium azide (NaN3), a scavenger of 1O228, was added to fixed A431 cells with and without the presence of EGF and the single-molecule efflux of H2O2 was again recorded. Compared to the un-stimulated control (green curve, Fig. 4h), NaN3 greatly diminished both the near and far-field portions of the H2O2 response to EGF (black and red curve, Fig. 4h). Further, we observed an increase in H2O2 after we exchanged water with D2O (the lifetime of 1O2 in D2O is 67 μs29, a factor of 16 times greater than that in water) for the EGF stimulation experiment on fixed A431 cell (purple curve, Fig. 4h), compared to the parallel experiment conducted with water (blue curve, Fig. 4h). In previous studies, the level of H2O2 before and after EGF stimulation is not affected when shutting down the mitochondria22–23. The fact that we were able to observe an obvious decrease in H2O2 level even below the basal level when adding NaN3 before and after EGF stimulation, and an increase in H2O2 level when extending the lifetime of 1O2, supports a complex pathway involving 1O2.
One possible signaling network that may explain this more complex response starts from , which is produced from the reduction of molecular oxygen by NAD(P)H oxidase in A431 cells (Fig. 4k)22, 26.
| (Reaction. 1) |
EGF was found to increase the production of in A431 cells while an inhibitor of NAD(P)H oxidase halts this mechanism in a manner that was also mitochondria independent22. This observation, together with the previous known EGF-EGFR induced NAD(P)H oxidase activation mechanism for EGF stimulated A431 cells from numerous literature6, 22–23, and our inhibition experiment results (Fig. S9a–b), supports Reaction 1. can then be decomposed by superoxide dismutase (SOD). It has been found that SOD is a master regulator of growth factor signaling and the inhibition on SOD1, which is an abundant copper/zinc enzyme found in the cytoplasm, increases the steady-state levels of and decreases the levels of H2O2 in A431 cells30. These reactions are rapid and occur with a diffusion limited reaction rate.
| (Reaction. 2) |
| (Reaction. 3) |
Transition metal ions such as iron or copper can catalyze the reduction of H2O2 by 33–35.
| (Reaction. 4) |
1O2 can decay to the ground state oxygen. It has been shown that the decay of 1O2 is determined by its interactions with water in the cell and not by interactions with other cell constituents with a decay rate constant of 3×105 s−1 36.
| (Reaction. 5) |
Ferrous also reacts with H2O2, known as the Fenton reaction37.
| (Reaction. 6) |
| (Reaction. 7) |
Upon EGF stimulation, 1O2 is converted into H2O2 catalyzed by EGF-EGFR38–39.
| (Reaction. 8) |
From solving the proposed pathway numerically (with the initial values for in vitro ROS obtained from the literature40: superoxide, ~1nM, H2O2, 1μM, 1O2, ~1nM) assuming a well-mixed condition and using the methods in previous work41–42, the concentration of and H2O2 increases with the addition of EGF (Fig. 4i), consistent with the experimental observations by us and others6, 17, 22. The addition of NaN3 causes decrease of H2O2 (Fig. 4j), even below the initial value, consistent with our observations in Fig. 4h. While the pathway that we proposed here is compelling, future work is necessary in order to conclusively rule out alternate mechanisms.
Conclusions
In conclusion, an array of SWNT sensors has been used to image, for the first time, the incident flux of H2O2 molecules that stochastically absorb and quench the emission with spatial and temporal resolution. Notably, arrays of this type can distinguish between molecules originating near an interface and those with no memory of origination, attributed as the far-field component. The signaling activity of EGFR on single A431 cells has been successfully measured using this sensor array. We find that the EGF stimulation induces on average 2 nmol H2O2 over a period of 50 min in A431 cells. Corresponding inhibition experiments suggest a mechanism whereby water oxidizes 1O2 at a catalytic site on the receptor itself, generating H2O2 in response to receptor binding. An EGFR-mediated H2O2 generation pathway that is consistent with all current and previous findings has been proposed and numerically tested for consistency.
Methods
Suspension of SWNT in Collagen
Single-walled HiPco carbon nanotubes (Rice University) were suspended in type 1 collagen (BD Biosciences) via 1min probe-tip sonication (1/4” tip, 40% amplitude). One mg of SWNT was used per mL of 3.41 mg/mL collagen stock in 0.02 N acetic acid for sonication. The mixture was centrifuged for 270min at 16300g and the pellet discarded, retaining the supernatant for future experiments.
Collagen-SWNT Thin Films
Collagen-SWNT was diluted with stock collagen (3.41 mg/mL) to make 272 mg/L SWNT concentration. This solution was diluted to 50 μg/mL collagen with 0.02 N acetic acid with a final concentration of SWNT of 8mg/L for imaging purposes (the concentration of SWNT is found by trial and error to achieve a desirable coverage of SWNT on the film) and pipetted onto glass bottom 35mm Petri dishes (MatTek Corp., P35G-1.5–14-C) in 500 μL aliquots to completely cover the glass region in the center of the dish. The collagen was dried at room temperature in a laminar flow hood. The dried film was rinsed well with PBS to remove the remaining acid. Everything was done under a sterilized environment.
Singlet Oxygen and Superoxide Generation
Rose bengal was used to generated singlet oxygen and superoxide in real time using a procedure described from a previous study43. Briefly, 50 nM of rose Bengal was illuminated at 561 nm at 200mW for 30 min and the fluorescence of nanotubes upon this illumination was recorded in real time. It is reported that this procedure will generate both singlet oxygen and superoxide. MnO2 was added to the solution to prevent any interference from H2O2.
Fluorescence Microscopy on Live and Fixed Cell
Human epidermoid carcinoma A431 cells and murine NIH-3T3 cells were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, Gemini Bio-Products) and 1% Pen-Strep Solution (10,000 U/mL Penicillin-G 10,000 μg/mL Streptomycin Penicillin-Streptomycin Solution, Gemini Bio-Products) at 37 °C with 5% CO2 on a collagen film10 in a glass bottom 35mm Petri dish (MatTek Corp., P35G-1.5–14-C) after serum starvation17. Right before imaging, the cell medium was changed into Leibovitz’s L-15 medium, which buffers the pH in the atmosphere. The nanotubes in the collagen film beneath the cells are then imaged using a fluorescence microscope (Carl Zeiss, Axiovert 200), with a CCD camera (Carl Zeiss, ZxioCam MRm) and 2D InGaAs array (Princeton Instruments OMA 2D). Movies were acquired using the WinSpec data acquisition program (Princeton Instruments). The nanotubes were excited by a 658 nm laser (LDM-OPT-A6–13, Newport Corp) at 35mW. After a stable fluorescence intensity was observed (Fig. S1), 500 μL of each reagent was added to reach the desirable final concentration. For experiment with fixed cells, A431 cells were washed with PBS, fixed in 4% paraformaldehyde (pH 7.4) for 10 min, washed 3 times and ready for imaging.
Fluorescent Staining of Cells
Cells were incubated with 4% PFA/PBS (USB Corporation) at 4 °C for 5 min, at room temperature for 10 min, then with 100% methanol (Sigma) at −20 °C for 10 min. The fixed cells were washed 3 times with PBS (Hyclone), permeabilize with 0.1% Triton X 100 (Sigma)/PBS for 20 min at room temperature followed by another washing with PBS. The cells were then incubated in 1% FBS/0.05% Tween (Sigma)-20/PBS for 20 min at room temperature, after which they were incubated with the primary antibody (rabbit polyclonal to EGFR, ABCAM Inc) in 1% FBS/0.05% Tween-20/PBS for 1h at room temperature. Washing and blocking were repeated. In the dark, the secondary antibody (Alexa Fluor 568 donkey anti-rabbit IgG, Invitrogen) was added in 1% FBS/0.05% Tween-20/PBS for 1h at room temperature (Alexa 1:500), after which 4’,6-diamidino-2-phenylindole (DAPI, Sigma Aldrich Co.) was added with a final concentration of 1μg/L. The washing step was repeated. The sample was then mounted in Moviol (Shandon Immu-Mount, Thermo Fisher Scientific). The samples were then analyzed in Zeiss LSM 510 Meta confocal microscope using the same configuration and processed in LSM image Browser software from Zeiss.
Atomic Force Microscope (AFM)
MFP-3D (Asylum Research) was used for tapping mode atomic force microscopy (AFM) imaging. Samples were directly deposited on a 75 mm × 25 mm glass slide (VWR International) and imaged using rectangular silicon tips (Olympus AC240TS) with a nominal spring constant of 2 N/m. Both topographic and height images were recorded during AFM analysis. Height analysis was performed using Igor Pro software.
Supplementary Material
Acknowledgements
M.S.S is grateful for a Beckman Young Investigator Award and a National Science Foundation (NSF) Career Award. This work was funded under the NSF Nanoscale Interdisciplinary Research Team on single-molecule detection in living cells using carbon nanotube optical probes. Part of this work was supported by the national grants Ministry of Education of the Czech Republic project No. MSM0021620806 and KAN Grant No. 400100701. We thank S. Tannenbaum, G.Wogan and L. Trudel and acknowledge a seed grant from the Center for Environmental Health Sciences at MIT. We also thank M. Balastik at Harvard Medical School for assistance with the confocal experiments, K. D. Wittrup, G. Stephanopoulos, J-H. Ahn, J-H Han at Chemical Engineering at MIT, S. Sheffield, Mathematics Department, MIT and Y. Li at University of Illinois Urbana Champaign for helpful discussions.
Footnotes
Additional Information
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/. Correspondence and requests for materials should be addressed to M.S.S..
References
- 1.Imlay JA Cellular defenses against superoxide and hydrogen peroxide. Annual Review Of Biochemistry 77, 755–776 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Veal EA, Day AM & Morgan BA Hydrogen peroxide sensing and signaling. Molecular Cell 26, 1–14 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Belousov VV et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods 3, 281–286 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Casanova D et al. Single europium-doped nanoparticles measure temporal pattern of reactive oxygen species production inside cells. Nat. Nanotechnol 4, 581–585 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Lee D et al. In vivo imaging of hydrogen peroxide with chemiluminescent nanoparticles. Nat. Mater 6, 765–769 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Miller EW, Tulyanthan O, Isacoff EY & Chang CJ Molecular imaging of hydrogen peroxide produced for cell signaling. Nat. Chem. Biol 3, 263–267 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Zhou M, Diwu Z, Panchuk-Voloshina N & Haugland RP A Stable Nonfluorescent Derivative of Resorufin for the Fluorometric Determination of Trace Hydrogen Peroxide: Applications in Detecting the Activity of Phagocyte NADPH Oxidase and Other Oxidases. Analytical Biochemistry 253, 162–168 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Hong Y, Blackman NMK, Kopp ND, Sen A & Velegol D Chemotaxis of nonbiological colloidal rods. Phys. Rev. Lett 99 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Cognet L et al. Stepwise quenching of exciton fluorescence in carbon nanotubes by single-molecule reactions. Science 316, 1465–1468 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Jin H, Heller DA, Kim J-H & Strano MS Stochastic Analysis of Stepwise Fluorescence Quenching Reactions on Single-Walled Carbon Nanotubes: Single Molecule Sensors. Nano Letters 8, 4299–4304 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Heller DA et al. Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes. Nature Nanotechnology 4, 114–120 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Yarden Y & Sliwkowski MX Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol 2, 127–137 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Herbst RS Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys 59, 21–26 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Lax I et al. Functional-Analysis of the Ligand-Binding Site of Egf-Receptor Utilizing Chimeric Chicken Human Receptor Molecules. Embo J. 8, 421–427 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masui H, Castro L & Mendelsohn J Consumption of Egf by A431 Cells - Evidence for Receptor Recycling. Journal of Cell Biology 120, 85–93 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter G & Cohen S Epidermal Growth-Factor. Annu. Rev. Biochem 48, 193–216 (1979). [DOI] [PubMed] [Google Scholar]
- 17.Bae YS et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide - Role in EGF receptor-mediated tyrosine phosphorylation. Journal of Biological Chemistry 272, 217–221 (1997). [PubMed] [Google Scholar]
- 18.Foote CS in. (ed. Pryor WA) 85–133 (Academic, New York; 1976). [Google Scholar]
- 19.Ziyatdinova GK, Gil’metdinova DM & Budnikov GK Reactions of superoxide anion radical with antioxidants and their use in voltammetry. J. Anal. Chem 60, 49–52 (2005). [Google Scholar]
- 20.Kim J-H et al. The rational design of nitric oxide selectivity in single-walled carbon nanotube near infrared fluorescence sensors for biological detection. Nature Chemistry 1, 473–481 (2009). [DOI] [PubMed] [Google Scholar]
- 21.DeYulia GJ, Carcamo JM, Borquez-Ojeda O, Shelton CC & Golde DW Hydrogen peroxide generated extracellularly by receptor-ligand interaction facilitates cell signaling. Proceedings of the National Academy of Sciences of the United States of America 102, 5044–5049 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morazzani M et al. Monolayer versus aggregate balance in survival process for EGF-induced apoptosis in A431 carcinoma cells: Implication of ROS-P38 mapk-integrin A2B1 pathway. Int. J. Cancer 110, 788–799 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Park HS et al. Sequential activation of phosphatidylinositol 3-kinase, beta Pix, Rac1, and Nox1 in growth factor-induced production of H2O2. Mol. Cell. Biol 24, 4384–4394 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramey NA, Park CY, Gehlbach PL & Chuck RS Imaging mitochondria in living corneal endothelial cells using autofluorescence microscopy. Photochem. Photobiol 83, 1325–1329 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Welsher K, Liu Z, Daranciang D & Dai H Selective probing and imaging of cells with single walled carbon nanotubes as near-infrared fluorescent molecules. Nano Letters 8, 586–590 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Nieva J & Wentworth P The antibody-catalyzed water oxidation pathway - a new chemical arm to immune defense? Trends Biochem.Sci 29, 274–278 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Carpenter G in The EGF Receptor Family: Biologic Mechanisms and Role in Cancer 33–60 (Academic Press, St. Louis; 2003). [Google Scholar]
- 28.Harbour JR & Issler SL Involvement of the Azide Radical in the Quenching of Singlet Oxygen by Azide Anion in Water. Journal of the American Chemical Society 104, 903–905 (1982). [Google Scholar]
- 29.Kuimova MK, Yahioglu G & Ogilby PR Singlet Oxygen in a Cell: Spatially Dependent Lifetimes and Quenching Rate Constants. Journal Of The American Chemical Society 131, 332–340 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Juarez JC et al. Superoxide dismutase 1 (SOD1) is essential for H2O2-mediated oxidation and inactivation of phosphatases in growth factor signaling. Proceedings of the National Academy of Sciences of the United States of America 105, 7147–7152 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JL, Wang LC, Chang CY & Liu TY Singlet oxygen is the major species participating in the induction of DNA strand breakage and 8-hydroxydeoxyguanosine adduct by lead acetate. Environ. Mol. Mutagen 33, 194–201 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Fridovich I Biology of Oxygen Radicals. Science 201, 875–880 (1978). [DOI] [PubMed] [Google Scholar]
- 33.Imlay JA, Chin SM & Linn S Toxic DNA Damage by Hydrogen-Peroxide through the Fenton Reaction Invivo and Invitro. Science 240, 640–642 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Halliwell B & Aruoma OI DNA Damage by Oxygen-Derived Species - Its Mechanism and Measurement in Mammalian Systems. FEBS Lett 281, 9–19 (1991). [DOI] [PubMed] [Google Scholar]
- 35.Khan AU & Kasha M Singlet Molecular-Oxygen in the Haber-Weiss Reaction. Proceedings of the National Academy of Sciences of the United States of America 91, 12365–12367 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatz S, Lambert JDC & Ogilby PR Measuring the lifetime of singlet oxygen in a single cell: addressing the issue of cell viability. Photochemical & Photobiological Sciences 6, 1106–1116 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Hardwick TJ The rate constant of the reaction between ferrous ions and hydrogen peroxide in acid solution. Canadian Journal of Chemistry 35, 428–436 (1957). [Google Scholar]
- 38.Wentworth P et al. Antibody catalysis of the oxidation of water. Science 293, 1806–1811 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Wentworth AD, Jones LH, Wentworth P, Janda KD & Lerner RA Antibodies have the intrinsic capacity to destroy antigens. Proceedings of the National Academy of Sciences of the United States of America 97, 10930–10935 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imlay JA Pathways of oxidative damage. Annu. Rev. Microbiol 57, 395–418 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Zanthoff H & Baerns M Oxidative coupling of methane in the gas phase. Kinetic simulation and experimental verification. Industrial & Engineering Chemistry Research 29, 2–10 (2002). [Google Scholar]
- 42.Zhu JY, Dittmeyer R & Hofmann H Application of sensitivity analysis to the reduction of a complex kinetic model for the homogeneous oxidative coupling of methane. Chemical Engineering and Processing 32, 167–176 (1993). [Google Scholar]
- 43.Mizukawa H & Okabe E Inhibition by singlet molecular oxygen of the vascular reactivity in rabbit mesenteric artery. British Journal Of Pharmacology 121, 63–70 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.