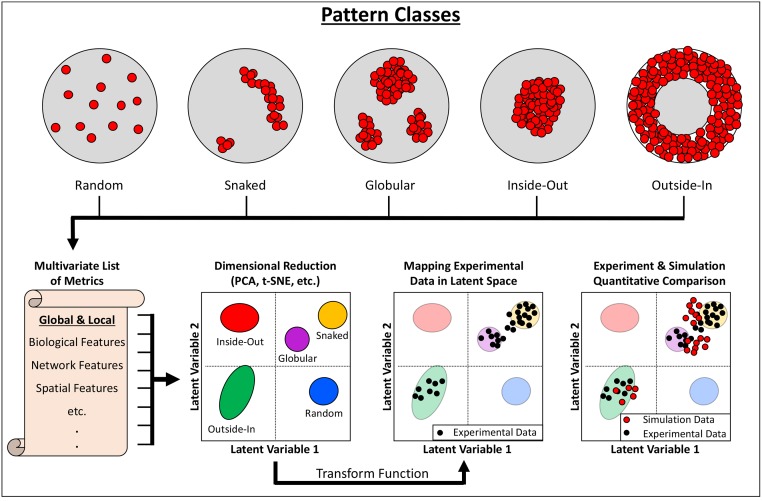
Fig 5. Overview of the analysis of spatial features using a combination of network analysis and dimensional reduction techniques.
A set of metrics is calculated or extracted from a series of pattern classes that depict typical cell organizations within the system—here, using the defined pattern classes from [133, 134]. The selected metrics should be equally represented in the simulated and experimental systems. Dimensional reduction techniques allow the multivariate data to be condensed to a few axes, ideally separating the defined pattern classes into distinct regions of the latent space. A transform function trained on the pattern class data can then be applied to metrics calculated from experimental and modeling results, mapping both into latent space and compared to the locations of the pattern classes. PCA, principal component analysis; t-SNE, t-distributed stochastic neighbor embedding.