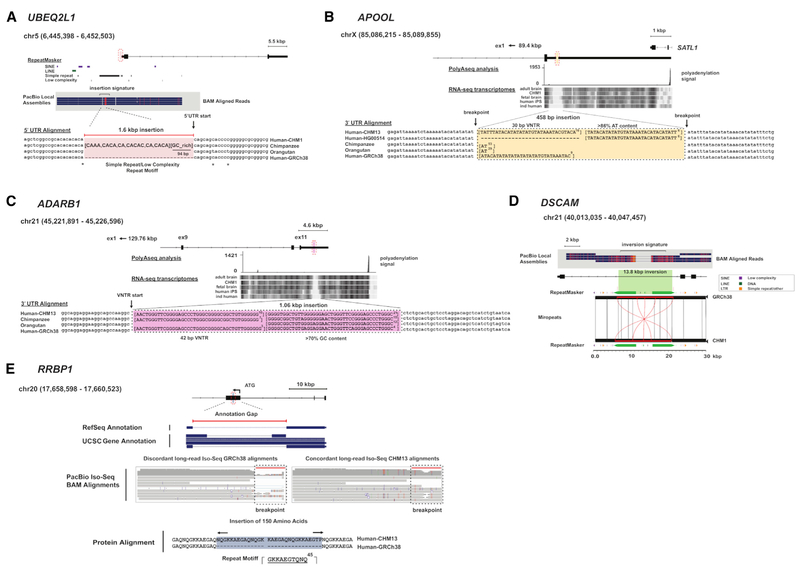
Figure 4. Missing Genic and Regulatory Sequence.
(A) A shared 1.6 kbp insertion in the 5’ UTR of UBEQ2L1 is almost completely comprised of simple repeat units (CACA) or low-complexity, GC-rich sequences. The breakpoints lie precisely at the start position of the 5’ UTR, and the missing sequence is largely conserved among chimpanzee and orangutan haplotypes.
(B) A 458 bp insertion is detected in 50% of the discovery samples in the large 5.63 kbp 3’ UTR of APOOL. The insertion is comprised of an AT-rich repeat array consisting of 30 bp units for a total of 24 tandem copies. Because of its AT-rich sequence composition, analysis with RNA-seq is inconclusive (“ind human” is a brain sample from a single anonymous individual). Comparison with nonhuman primates reveals that the repeat array is largely absent.
(C) A 1.1 kbp shared insertion in the 3’ UTR of the ADARB1 corresponds to a large VNTR comprised primarily of GC-rich sequence. Each repeat unit is 42 bp with a variable number of copies present in CHM13, chimpanzee, and orangutan. We detect 31 tandem copies in CHM13 compared to only 7 in the GRCh38 reference assembly.
(D) A 13.8 kbp inversion in intron 32 of DSCAM. The shared inversion is flanked by inverted, complete LINE-L1 repeat sequences.
(E) A 480 bp shared insertion detected in the first exon of RRBP1 (transcript ENST00000246043.8) is associated with gaps in RefSeq and UCSC gene annotations (top). Mapping human IPS-derived PacBio Iso-Seq data to the GRCh38 reference assembly identifies discordant read alignments at the insertion site (Iso-Seq alignments, left). Analysis of the insertion and adjacent flanking sequence identifies a large VNTR (1,380 bp) comprised of 30 bp repeat units. In our discovery set, the number of copies varies between 15 (450 bp) and 16 (480 bp). Translation of the newly assembled haplotype sequence from CHM13 (15 copies, 450 bp) shows that the insertion maintains the open reading frame and adds an additional 150 amino acids (Iso-Seq alignments right).
For each panel: regions of shared or major allele structural variation are annotated and compared between GRCh38, alternate human reference assemblies (CHM1/CHM13), and nonhuman primates. Multiple sequence alignments were generated using MAFFT or visualized using Miropeats against sequenced large-insert clones. Additional functional annotations are shown using short-read Illumina RNA-seq data, PolyA-seq, and PacBio long-read Iso-Seq data.