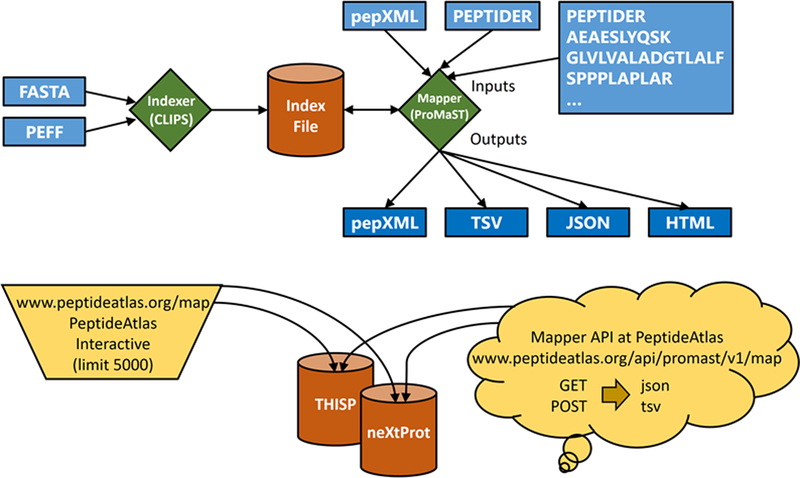
Figure 1:
Graphical overview of all ProteoMapper components. The indexer takes as input a FASTA or PEFF file and creates a segmented index. The mapper takes as input one or more peptides as a list or a pepXML file, a previously created index, and other optional parameters. It then provides its output in pepXML, TSV, JSON, or HTML. In addition to a downloadable form for local use, an interactive web page and a web service are available for a predefined set of reference proteomes at PeptideAtlas.