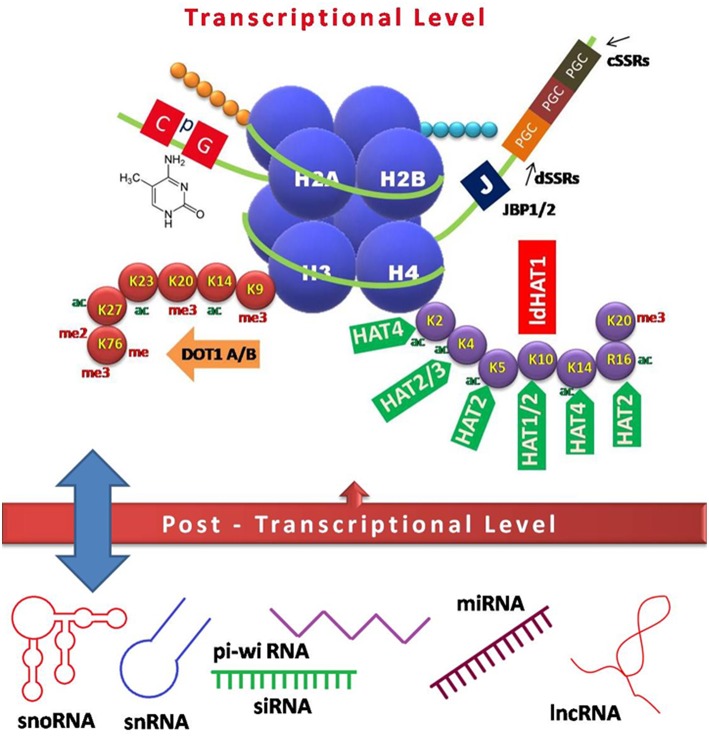
Figure 1.
Schematic model of epigenetic regulation during Leishmania infection. Interplay of various factors involved in controlling gene expression at the transcriptional (DNA and histone modifications) and post-transcriptional level (non-coding RNAs) is depicted. CpG-rich regions of repressed genes are usually methylated, which in turn recruit chromatin modifiers to keep the genes in any of the three states, i.e., repressed, expressed, or poised. Heavily expressed genes show neither DNA methylation nor acetylated histones, while repressed genes tend to have both methylated DNA and histones, which inhibit the accessibility of polymerases, and other factors required for transcription. Base J, a DNA modification is crucial for transcriptional control in Leishmania species. Various non-coding RNAs arising mostly from UTRs act as regulatory elements in a feedback loop (13, 14). JBP, Base J binding protein; PGC, polycistronic gene cluster; cSSRs, convergent strand switch regions; dSSRs, divergent strand switch regions; DOT, disrupter of telomere; ac, acetylation; me, methylation; me2, dimethylation; me3, trimethylation; HAT, histone acetyl transferase; sno, small nucleolar; sn, small nuclear; pi-wi, piwi interacting; si, small interfering; mi, micro; lnc, long non-coding.