Figure 3.
Endocytosis Alteration in Dnm2R465W/+ KI Myoblasts Is Rescued by Allele-Specific Inactivation or Correction
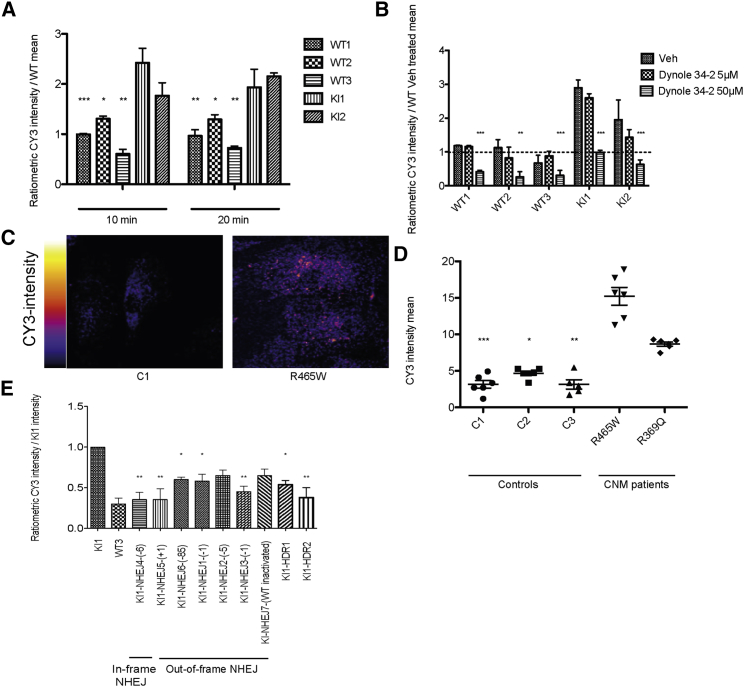
(A) Transferrin uptake assays in three WT and two Dnm2R465W/+ KI mouse myoblast lines after a 10 or 20 min incubation with CY3-labeled transferrin, normalized to the mean CY3 intensity of the WT1 cells. CY3 mean geometric fluorescence intensity was measured by FACS from three independent experiments. Data are expressed as the mean ± SEM. One-way ANOVA followed by Dunnett’s multiple-comparison test. *p < 0.05, **p < 0.01, ***p < 0.001. (B) Transferrin uptake in three WT and two Dnm2R465W/+ KI mouse myoblast lines after a 10 min incubation upon treatment with the dynamin inhibitor Dynole 34-2 used at two different concentrations in DMSO. CY3 mean geometric fluorescence intensity was measured by FACS from three independent experiments and normalized to the mean of WT DMSO-treated cells (represented by the dotted line). Data are expressed as the mean ± SEM. One-way ANOVA followed by Dunnett’s multiple-comparison test. **p < 0.01, ***p < 0.001 versus DMSO-treated cells. (C) Primary human myoblasts from healthy or CNM patients with the R465W mutation were incubated with CY3-transferrin for 10 min and imaged by confocal microscopy. (D) Quantification of CY3 intensity of three independent experiments in duplicate, as in (C). Three WT myoblast lines and two patient myoblast lines with either an R465W or an R369Q heterozygous mutation were compared. Controls 1 and 2 and the R465W patient cells are primary myoblasts, whereas control 3 and the R369Q patient cells are immortalized myoblasts. Quantified were 125–217 cells per line. Data are expressed as the mean ± SEM. Non-parametric Kruskal-Wallis and post hoc Dunn’s multiple-comparison tests. *p < 0.05, **p < 0.01, ***p < 0.001 versus R465W. (E) Quantification of three independent experiments as in (A). Transferrin uptake assays in one WT and one Dnm2R465W/+ KI mouse myoblast line, seven NHEJ-derived clones, and two HDR-derived clones after 10 min incubation with CY3-labeled transferrin, normalized to the CY3 intensity of the KI1 cells. Indels are indicated in the number of nucleotides in parentheses. One in-frame NHEJ and six out-of-frame NHEJ clones were used. The KI1-NHEJ7 clone had an out-of-frame indel specifically on the WT allele. Data are expressed as the mean ± SEM. One-way ANOVA followed by Dunnett’s multiple-comparison test. *p < 0.05, **p < 0.01 versus KI1. No statistically significant difference compared to WT3.