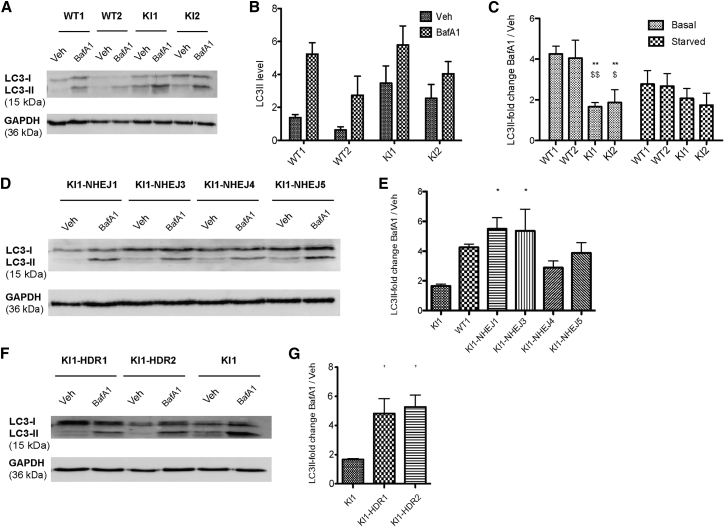
Figure 4.
Autophagy Defects in Dnm2R465W/+ KI Myoblasts Are Rescued by Allele-Specific Inactivation or Correction
(A) WT and KI immortalized myoblasts were treated or not with bafilomycin A1 (BafA1 in DMSO) under basal conditions, and lipidated LC3-II was detected by western blot. GAPDH was used as the loading control. (B) Quantification of LC3-II levels in WT and KI myoblasts under basal conditions without any treatment. Four independent experiments. Data are expressed as the mean ± SEM. One-way ANOVA followed by Bonferroni’s multiple-comparison test showed no statistically significant difference between WT and KI myoblasts. (C) Quantification of BafA1-induced accumulation of LC3-II relative to GAPDH in two WT and two KI myoblast lines under basal or starved conditions, expressed as the ratio of BafA1-treated cells versus untreated cells. Four independent experiments. Data are expressed as the mean ± SEM. One-way ANOVA followed by Bonferroni’s multiple-comparison test. **p < 0.01 versus WT1; $p < 0.05, $$p < 0.01 versus WT2. (D) Levels of LC3-II relative to GAPDH in allele-specific NHEJ inactivated myoblasts treated or not with BafA1 under basal conditions. Clone KI1-NHEJ4 has an in-frame deletion in Dnm2, whereas the other clones carry out-of-frame indels. GAPDH was used as the loading control. (E) Quantification of BafA1-induced accumulation of LC3-II relative to GAPDH in four allele-specific NHEJ inactivated myoblast lines under basal conditions, expressed as the ratio of BafA1-treated cells versus untreated cells. Clone KI1-NHEJ4 has an in-frame deletion in Dnm2, whereas the other clones carry out-of-frame indels. Three independent experiments. Ratio in the WT1 and KI1 myoblasts from (C) are shown as a comparison. Data are expressed as the mean ± SEM. One-way ANOVA followed by Bonferroni’s multiple-comparison test. *p < 0.05 versus KI1. No statistically significant difference compared with WT1. (F) Levels of LC3-II in two allele-specific HDR-corrected myoblast lines and the non-edited KI1 myoblasts, treated or not with BafA1, under basal conditions. GAPDH was used as the loading control. (G) Quantification of BafA1-induced accumulation of LC3-II relative to GAPDH in two allele-specific HDR-corrected myoblast lines and the non-edited KI1 myoblasts under basal conditions, expressed as ratio of BafA1-treated cells versus untreated cells. Three independent experiments. Data are expressed as the mean ± SEM. One-way ANOVA followed by Bonferroni’s multiple-comparison test. *p < 0.05 versus KI1.