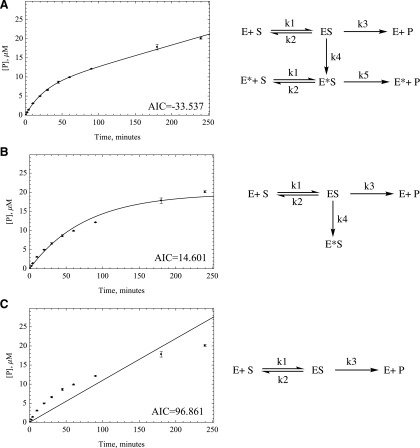
Fig. 4.
Two different kinetic models were developed to compare the numerical fitting of time-course data with MM. (A) MM model, (B) dead enzyme model, (C) MAM. Akaike values were provided by Mathematica to compare the goodness of fit, proving that MAM is the best model to fit the time-course data for most substrates except for zoniporide. Data- fitting plots for O6BG are provided next to each kinetic scheme for comparison. The Akaike values for O6BG are provided on the bottom right side of each plot.