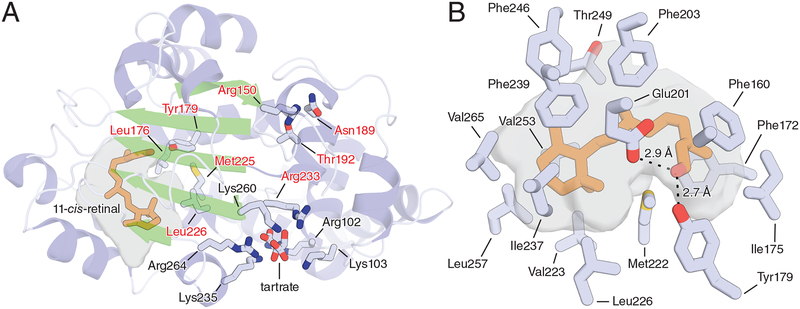
Figure 5. Structural basis of 11-cis-retinoid binding by CRALBP.
A) Overall structure of wild-type human CRALBP. The cavities housing the bound 11-cis-retinaldehyde (orange sticks) are shown as a grey surface. Residues known to be mutated in retinal diseases are labeled with red text. Residues labeled in black (as well as Arg233) make up a basic patch on the protein surface that is thought to bind acidic phospholipids to facilitate retinoid release from the protein. The structure features a bound tartrate molecule at the center of the basic patch which possibly mimics the structure of an acidic phospholipid head group. B) Close-up view of the retinoid-binding cavity of CRALBP showing residues responsible for forming the bend geometry of the cavity as well as those involved in dipolar interactions with the terminal aldehyde moiety. Residue numbering refers to the mature protein lacking the initiating Met residue.