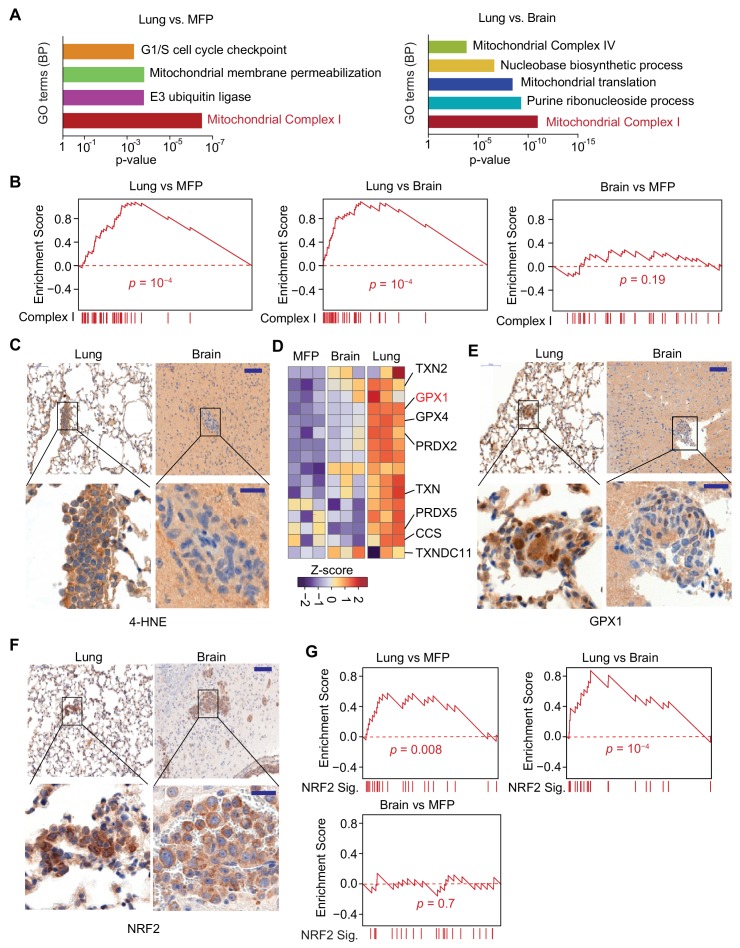
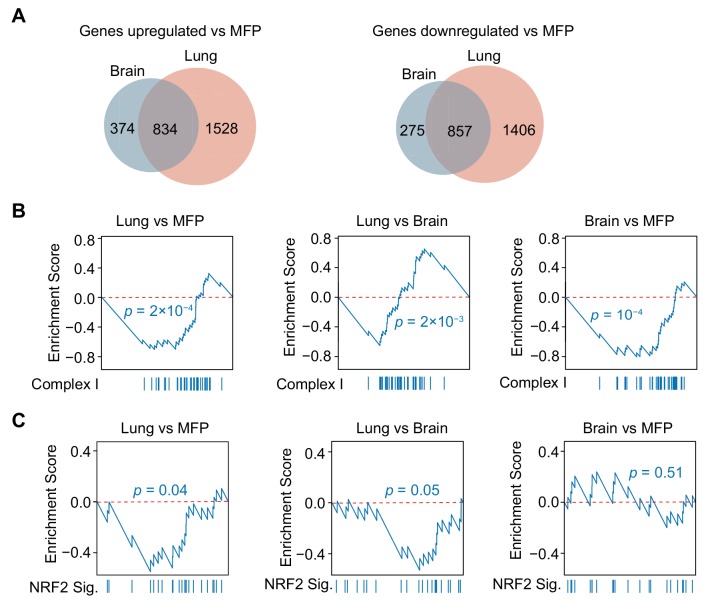
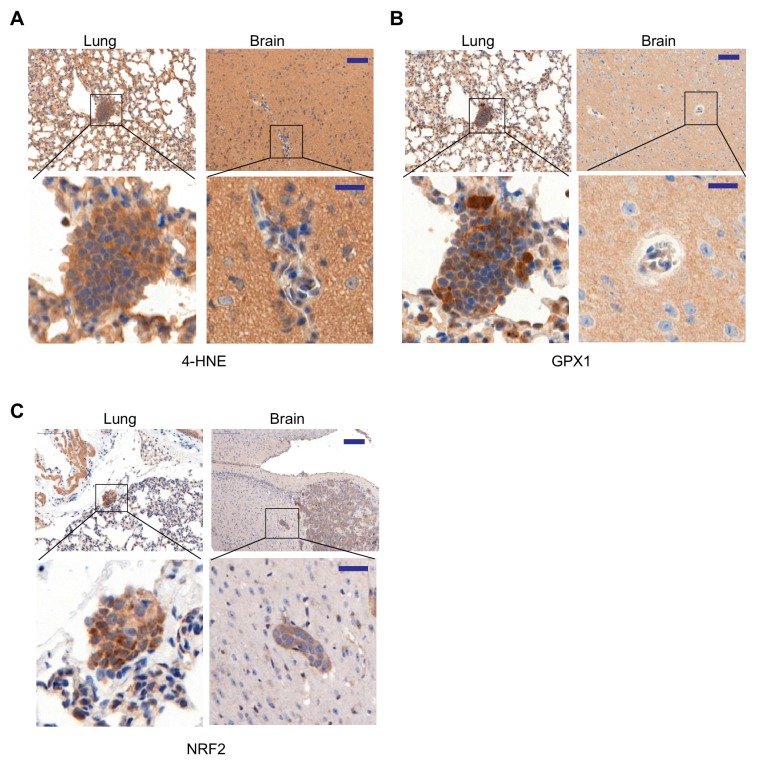
Figure 5. Mitochondrial Complex I expression and oxidative stress in lung micrometastases.
(A) Gene Ontology (GO) analysis of biological processes (BP) of genes that were upregulated in MDA231 lung micrometastases compared to brain micrometastases or mammary tumors. The top functional groups and their corresponding pvalues are shown (n = 3); (B) Gene Set Enrichment Analysis (GSEA) analysis of nuclear Complex I genes was performed for the genes identified by Flura-seq in the indicated pairs of MDA231 lung and brain micrometastases and mammary tumors. p-Values were calculated by random permutations; (C) Oxidative stress in lung and brain tissue sections containing micrometastases were examined by IHC using anti-4-HNE antibody. Scale bars, 100 μm (top) and 20 μm (bottom); (D) Heatmap representation of the expression of genes encoding known antioxidant factors in MDA231 tumors from the indicated organs. The highlighted genes were also upregulated in clinical samples of lung metastasis from breast cancer patients (Figure 6D); (E) IHC analysis of GPX1, an antioxidant gene product identified by Flura-seq to be selectively upregulated in lung micrometastases. Scale bars, 100 μm (top) and 20 μm (bottom); (F) IHC analysis of NRF2 in lung and brain micrometastases. Scale bars, 100 μm (top) and 20 μm (bottom); (G) GSEA analysis of the NRF2 response gene signature applied to Flura-seq data from the indicated pairs of MDA231 lung and brain micrometastases and mammary tumors (n = 3). p-Values were calculated by random permutations.