Polymyxins, including polymyxin B and polymyxin E (colistin), are now increasingly being used worldwide to treat patients with multidrug-resistant (MDR) Gram-negative bacterial infections. This necessitates that laboratories employ an accurate and reliable method for the routine performance of polymyxin susceptibility testing.
KEYWORDS: antimicrobial susceptibility, colistin, Gram-negative bacteria, polymyxin resistance
ABSTRACT
Polymyxins, including polymyxin B and polymyxin E (colistin), are now increasingly being used worldwide to treat patients with multidrug-resistant (MDR) Gram-negative bacterial infections. This necessitates that laboratories employ an accurate and reliable method for the routine performance of polymyxin susceptibility testing. A number of reasons have accounted for the difficulties with susceptibility testing for the polymyxins, including their multicomponent composition, poor diffusion in the agar medium, adsorption to microtiter plates, the lack of a reliable susceptibility test, the lack of a specific breakpoint from professional organizations, the synergistic effect of polysorbate 80, and the development of heteroresistance. This minireview discusses such problems that impact the results of currently available susceptibility testing methods. We also provide emerging concepts on mechanisms of polymyxin resistance, including chromosomally and plasmid-mediated mcr-related resistance. Broad-range investigations on such critical issues in relation to polymyxins can be beneficial for the implementation of effective treatment against MDR Gram-negative bacterial infections.
INTRODUCTION
The increase in antimicrobial resistance has currently become a serious threat to public health worldwide, having a significant impact on infection control practices. If this trend continues, simple infections will no longer be treatable (1, 2). This is particularly true for carbapenems, which used to be the first-line antimicrobials but which have been increasingly compromised and which no longer constitute salvage therapy against the Gram-negative bacilli isolated in hospitals (3–6).
POLYMYXINS
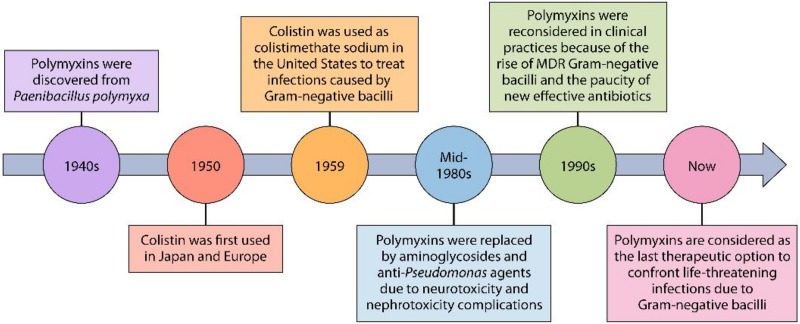
Polymyxins B and E (also known as colistin) are cyclic polypeptide antibiotics that are synthesized by nonribosomal enzymes by members of the Paenibacillus genus (7, 8). Polymyxin B was discovered in 1947 from Paenibacillus polymyxa, whereas colistin was produced by the growth of P. polymyxa subsp. colistinus in 1949 (9) (Fig. 1). These antibiotics were widely used to treat serious infections caused by Gram-negative bacilli until the mid-1980s, when they were banned in clinical practices because of their nephrotoxicity and neurotoxicity adverse events and also the availability of less toxic drugs, mainly antipseudomonal aminoglycosides (10–12). The rise of multiresistant Gram-negative bacilli and also the paucity of new effective antibiotics led to the reemergence of polymyxins as a last-resort treatment option by the mid-1990s (13, 14). Polymyxins are now a key part of the antibiotic armamentarium and serve as the last therapeutic option to confront life-threatening infections caused by multidrug-resistant (MDR) and extensively drug-resistant (XDR) pathogens, including Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae (15–18). Such antibiotic-resistant microorganisms are described by world authorities as “nightmare bacteria” that pose a “catastrophic threat” to people worldwide (19). They are also so important that the mortality rate in individuals infected with these bacteria can be as high as over 50% (17, 19).
FIG 1.
A polymyxin timeline from 1940s to today. Polymyxin B was discovered in 1947, whereas colistin was produced in 1949. Colistin has been available since 1959 for the treatment of infectious diseases caused by Gram-negative bacteria. Polymyxins were gradually abandoned in most parts of the word in about 1980. In recent years, polymyxins have been reconsidered in clinical practices for use against MDR strains of bacteria.
Chemical Structure.
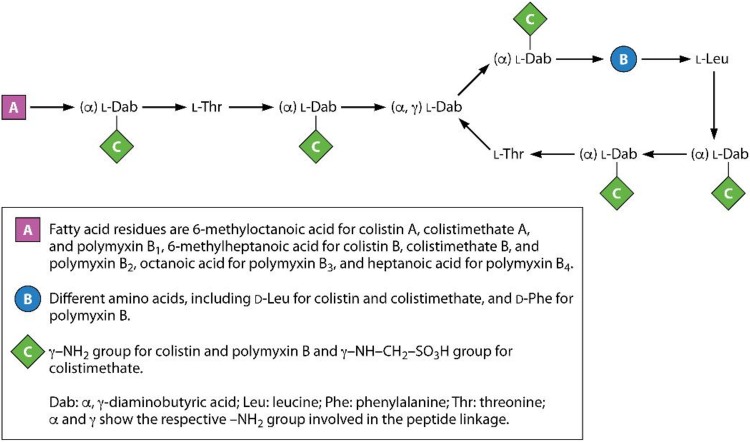
Polymyxins are polycationic peptides consisting of a cyclic heptapeptide, a linear tripeptide segment, and a fatty acid tail linked to the N terminus of the tripeptide (Fig. 2) (7, 8). They consist of both hydrophilic and hydrophobic regions that are essential for their antimicrobial activity. Two commercially available polymyxins, polymyxin B and polymyxin E (colistin), are very similar structurally. They differ by only a single amino acid in the heptapeptide ring, with a phenylalanine in polymyxin B and a leucine in colistin (7, 8). In addition, at least four major components of polymyxin B, B1, B2, B3, and B4, and two major types of colistin, A and B (also known as polymyxins E1 and E2, respectively), have been described, differing only in the structure of their N-terminal fatty acyl moiety (7, 8, 20).
FIG 2.
Structures of colistin, polymyxin B, and colistimethate.
Polymyxin B is administered parenterally, orally, and topically as sulfate salts. Two different forms of colistin are available for clinical practices: colistin sulfate and colistin methanesulfonate (CMS; or colistimethate). Colistin sulfate is used topically and orally, while CMS is administered by the parenteral and intravenous routes and by nebulization. CMS is less toxic than colistin sulfate when it is used parenterally (21). Indeed, CMS is formed by the reaction of colistin with formaldehyde and sodium bisulfite (22), leading to the addition of a sulfomethyl group to the primary amines of colistin. It is an inactive prodrug form of colistin that undergoes hydrolysis for conversion to the active compound, colistin, as well as a complex mixture of partially sulfomethylated derivatives in both in vitro aqueous media and in vivo biological fluids, taking several hours to achieve effective colistin concentrations (22–24).
Chemical Stability.
Importantly, the long-term storage of polymyxins can serve as a source of loss of potency and can provide untoward physicochemical modifications (25). Neither the Clinical and Laboratory Standards Institute (CLSI) nor the European Committee on Antimicrobial Susceptibility Testing (EUCAST) addressed the stability of or storage time for polymyxins in aqueous solutions. Polymyxin B was used as a component of a permeabilization reagent reported to be stable in aqueous solution for at least 2 months at 4°C (2 to 8°C) and more than 3 weeks at room temperature (26). Polymyxin B was found to be stable for at least 1 day when stored at 4 or 25°C in infusion bags containing 0.9% sodium chloride injection (pH 5.6). Stability did not differ significantly between the two storage temperatures (27). In addition, Li et al. (24) demonstrated in a study that solutions of colistin sulfate were very stable in water at 4°C. After 60 days, the percentages of colistin A (polymyxin E1) and B (polymyxin E2) remaining were 97.4% and 105.3%, respectively. In addition, there were no decreases in the concentrations of colistins A and B stored in water at 37°C for up to 120 h. However, they, especially colistin A, were less stable in isotonic phosphate buffer (pH 7.4) and human plasma at 37°C.
Mechanism of Action.
Due to the nature of the positive charge of polymyxins, they bind to the negatively charged phosphate groups of membrane lipids of Gram-negative bacilli. This displaces divalent Ca2+ and Mg2+ cations and destabilizes the lipopolysaccharide (LPS), consequently changing the permeability of the bacterial cell membranes, leading to leakage of the cytoplasmic content, and eventually causing cell death (8, 13, 28). It has been also demonstrated that polymyxins inhibit vital respiratory enzymes, such as type II NADH-quinone oxidoreductases in the bacterial cell membrane (29).
Spectrum of Activity.
Polymyxins have a narrow antibacterial spectrum, mainly most clinically significant Gram-negative bacilli, including members of the Enterobacteriaceae (Citrobacter spp., Enterobacter spp., Escherichia coli, Klebsiella spp., Salmonella spp., and Shigella spp.) and nonfermentative bacteria (Acinetobacter spp., P. aeruginosa, and most strains of Stenotrophomonas maltophilia) (12, 30–33). As described above, polymyxins have now become the last-line choice against infections caused by multiresistant Gram-negative bacilli, especially K. pneumoniae, P. aeruginosa, and A. baumannii (34–36). They are not active against Gram-negative cocci (Neisseria spp.), Gram-positive organisms, and most anaerobic bacteria (12, 30).
Mechanisms of Resistance.
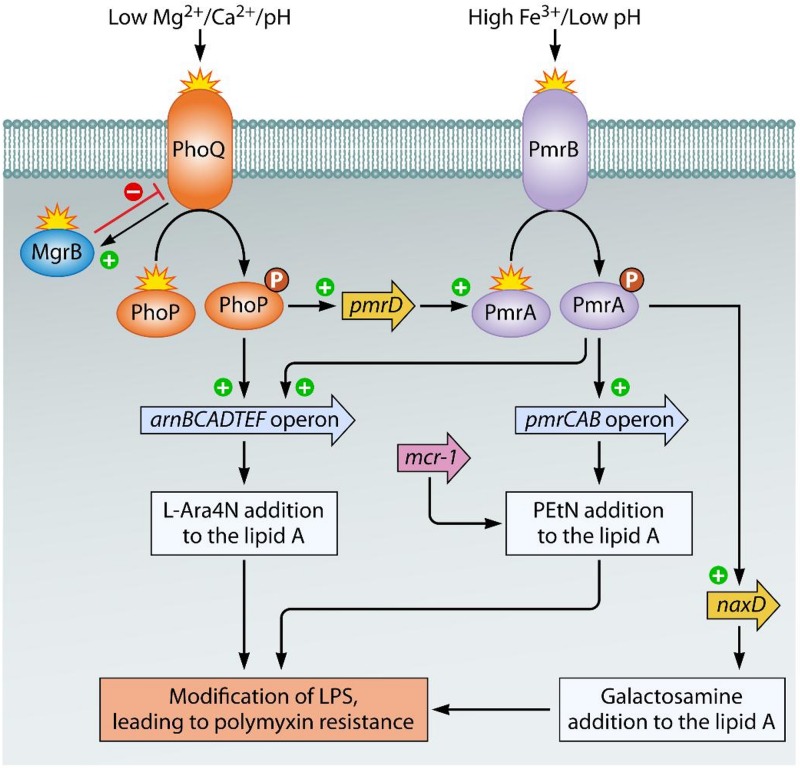
Fortunately, the resistance of Gram-negative bacilli to the polymyxins is yet uncommon. However, there are a number of reports of infections due to polymyxin-resistant bacteria that were normally susceptible to these drugs (5, 37–39). Modification of the LPS molecules is the primary mechanism responsible for resistance to polymyxins in Gram-negative bacteria (Fig. 3) (40–43). This resistance mechanism is as a result of chromosomal mutations and occurs frequently by the synthesis, transmembrane transport, and attachment of 4-amino-4-deoxy-l-arabinose (l-Ara4N) and/or phosphoethanolamine (PEtN) to a phosphate group in lipid A. Subsequently, the increased net charge of modified lipid A reduces the affinity of positively charged polymyxins, leading to resistance (44, 45). Attachment of l-Ara4N and/or PEtN units to the lipid A moiety of LPS has been identified in polymyxin-resistant Gram-negative bacilli, including Salmonella enterica serotype Typhimurium, Escherichia coli, K. pneumoniae, and P. aeruginosa (46–50). However, A. baumannii possesses no genetic apparatus necessary for the biosynthesis and transport of l-Ara4N; therefore, in addition to modification of lipid A with PEtN, glycosylation of the phosphate group of lipid A with d-galactosamine units is also associated with resistance to polymyxins (51, 52).
FIG 3.
Mechanisms of LPS modification involved in polymyxin resistance in Gram-negative bacilli. In Salmonella spp., Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa, the sensing of various stress conditions, such as the presence of cationic compounds (polymyxins), low Mg2+ and Ca2+ concentrations, acidic pH, and high Fe3+ concentrations, by the histidine kinases PhoQ and PmrB activates the two-component systems (TCSs) PhoP-PhoQ and PmrA-PmrB, respectively. Subsequent activation of the arnBCADTEF and pmrCAB operons leads to the synthesis and addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) and phosphoethanolamine (PEtN) to lipid A, respectively. In addition, PmrAB is activated by PhoP-PhoQ via the product of the pmrD gene, which in turn activates pmrA for activation of the arnBCADTEF operon. Inactivation of MgrB (a negative regulator of the PhoP-PhoQ system) by amino acid substitutions leads to overexpression of the phoP-phoQ operon as well, causing activation of the pmrHFIJKLM operon, thus leading to the production of l-Ara4N. On the other hand, polymyxin resistance in Acinetobacter baumannii is due to alterations in PmrB or the response regulator PmrA, which activates the PmrAB TCS. The subsequent upregulation of the pmrCAB operon and the naxD gene promotes the addition of PEtN and galactosamine to lipid A, respectively. In addition, a recently identified mcr-1 gene, encoding phosphoethanolamine transferase, has been shown to be the main mechanism of polymyxin resistance.
Two-component systems (TCSs), mainly PhoP-PhoQ and PmrA-PmrB, regulate the expression of most of the genes involved in LPS modification in Gram-negative bacilli. In Salmonella spp., E. coli, K. pneumoniae, and P. aeruginosa, activation of these TCSs under specific stress conditions generally leads to LPS modification. The inner membrane sensor histidine kinase PhoQ and/or PmrB is activated under conditions of bacterial growth in the presence of low concentrations of Mg2+ and Ca2+ and high concentrations of Fe3+ or in the presence of cationic compounds, including polymyxins. When sensor kinases (PhoQ, PmrB) are activated, they phosphorylate their cognate cytoplasmic response regulator (PhoP, PmrA), which in turn modulates the expression of the target genes arnBCADTEF (also called pmrHFIJKLM) and pmrCAB, responsible for LPS modifications by l-Ara4N and PEtN, respectively. Furthermore, constitutive activation of the systems of enteric bacteria and P. aeruginosa can be caused by missense mutations, leading to the upregulation of the corresponding operons, resulting in LPS modification (53). The histidine kinase pmrB seems to be a more common site for mutations than the response regulator gene pmrA. Polymyxin resistance in A. baumannii is primarily due to missense or small insertion/deletion mutations in pmrA and/or pmrB genes (51, 53). Similar to the findings for enteric bacteria, pmrB appears to be the most commonly mutated gene in A. baumannii. The complete loss of lipid A or the LPS core by mutations in the first three lipid A biosynthesis genes, lpxA, lpxB, and lpxC, and in the lpsB gene, encoding aglycosyltransferase (involved in the biosynthesis of the LPS core), has also been implicated in colistin resistance in A. baumannii (54).
A distinct resistance phenomenon, namely, heteroresistance, has appeared as a new challenge faced by antimicrobial therapy. This type of resistance to polymyxins has been described in several Gram-negative pathogens, including A. baumannii (55, 56), K. pneumoniae (57), Salmonella spp. (58), Enterobacter spp. (59), as well as P. aeruginosa (60). Heteroresistance is defined as resistance to certain antibiotics expressed by a subset of isolates of a microbe that are generally categorized as susceptible based on in vitro susceptibility tests (61). Heterogeneous colistin-resistant A. baumannii is known as a subpopulation of colistin-susceptible isolates with MICs in the range of 0.25 to 2 μg/ml and with the ability to grow in the presence of colistin at 3 to 10 μg/ml (62). These resistant subpopulations can be developed into highly or completely resistant isolates following colistin administration (63). In such cases, antibiotic therapy leads to elimination of the sensitive members of the bacterial subpopulation and, it is speculated, the selection of more resistant cells, which would, consequently, impede the therapeutic efficiency of antibiotics. Although the underlying molecular mechanism is unknown, data suggest that heteroresistance may occur as a result of mutations within the phoPQ or mgrB regulatory system (57, 64), as well as the complete loss of LPS production due to a mutation in or the insertional inactivation of lipid A biosynthesis genes lpxA, lpxC, and lpxD (54, 65). It is important to note that the heterogeneity of resistance to antibiotics is different from bacterial persistence, which was first described as the capacity of bacteria to tolerate exposure to lethal concentrations of penicillin (66). Persisters are mutant cells that neither die nor grow in the presence of an antibiotic, suggesting that they are in a state of dormancy (67), and they grow only after removal of the antibiotic. Additionally, the progeny of persisters does not display increased resistance to the antibiotic but shows the same pattern of susceptibility to the antibiotic as the original bacterial population (68).
Liu et al. discovered for the first time, in November 2015, transmissible polymyxin resistance in an E. coli isolate recovered from food animals and raw meat in China (69). The plasmid-borne gene mcr-1, which mediates the addition of PEtN moiety to lipid A (69), has since been described in different species of the family Enterobacteriaceae, such as K. pneumoniae, Shigella sonnei, Salmonella spp., and Enterobacter spp., from patients, animals, and environmental samples in different regions of the world (70–72). In a short time, 13 mcr-1 subgroups (mcr-1.1 to mcr-1.13) were identified in various species of Enterobacteriaceae in different countries (73). To date, eight mcr variants (mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, mcr-6, mcr-7, and mcr-8) showing nucleotide sequence identity to each other have been described in Enterobacteriaceae and other genera (73–77). Notably, a colistin-nonsusceptible P. aeruginosa strain carrying a chromosomal copy of the mcr-5 gene which was embedded within a Tn3-family transposon has recently been reported (78). This new mechanism confers a degree of resistance lower than that conferred by the chromosomally mediated mechanism of polymyxin resistance (79). However, this is a major global issue, where such transferable mechanisms are responsible for the rapid and ongoing dissemination of polymyxin resistance among clinical pathogens.
CHALLENGES IN POLYMYXIN SUSCEPTIBILITY TESTING
Due to the growing menace of multiresistant pathogens (80) and, consequently, the increased need for polymyxin treatment in critically ill patients with bacteremia and ventilator-associated pneumonia (9), a significant effort is required to maintain the antibacterial properties of these antibiotics. Optimization and standardization of in vitro polymyxin susceptibility testing and definition of the correct breakpoints are critical issues for both patient care and epidemiological surveillance purposes. In other words, because of their increased clinical use, the development of an accurate and reliable method for determining the susceptibility of isolates to polymyxins is now an urgent need. There are several issues associated with in vitro susceptibility testing of polymyxins, such as interpretive categories (criteria) and breakpoints, quality control (QC) strains, the reliable reference susceptibility testing method, the multicomponent composition of polymyxins, their poor diffusion into agar, their cationic nature, the effect of polysorbate 80 (P-80), and the development of heteroresistance, as well as the impact of the medium, subcultures, and storage (81). This minireview attempts to describe the different challenges, technical issues, and recommendations associated with polymyxin susceptibility testing, with the aim of guiding antimicrobial therapy.
Interpretive Criteria/Categories and Quality Control Strains.
There is no consensus between the two professional organizations establishing breakpoints for polymyxins, the Clinical and Laboratory Standards Institute (CLSI) (82) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (83) (Table 1), although attempts have recently been made to coordinate these. Neither CLSI nor EUCAST has recommended the disk diffusion (DD) method for polymyxin susceptibility testing, so there are currently no established zone diameter breakpoints for colistin or polymyxin B (82–84). Furthermore, MIC breakpoints for colistin and polymyxin B are available in CLSI guidelines for Pseudomonas spp. and Acinetobacter spp. but not for the Enterobacteriaceae, and also, no interpretative breakpoint for polymyxin B has been established by EUCAST guidelines (82, 83). CLSI has established a MIC epidemiological cutoff value (ECV) for colistin applicable to five members of the Enterobacteriaceae, including E. coli, K. pneumoniae, Klebsiella aerogenes (formerly Enterobacter aerogenes), Enterobacter cloacae, and Raoultella ornithinolytica (82). The ECV is generally the MIC or zone diameter value that divides bacterial populations into those with and those without phenotypically detectable resistance (non-wild-type [NWT] and wild-type [WT], respectively). An ECV for WT strains describes the isolates with no detectable resistance (due to acquired and/or mutational mechanisms) or reduced susceptibility toward the antimicrobial agent tested, while the ECV for NWT strains defines isolates with detectable resistance and reduced susceptibility to the antimicrobial agent being evaluated. It is noteworthy that ECVs are based on in vitro data only, using MIC or zone diameter distributions, and must not be used as clinical breakpoints of susceptible, intermediate, or resistant (82). Here, if the colistin MIC for the above-mentioned members of the Enterobacteriaceae is at or below the ECV (≤2 μg/ml), it can be assumed that the isolate is a WT strain. If the colistin MIC is ≥4 μg/ml, the isolate should be retested, and if the result is confirmed, the isolate can be considered NWT.
TABLE 1.
Polymyxin B and colistin interpretative breakpoints according to CLSI and EUCAST guidelines in 2018
| Microorganism | Polymyxin | CLSIa
,b
|
EUCASTa
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disk content | Zone diameter interpretative criteria (mm) |
MIC interpretative criteria (μg/ml) |
Disk content | Zone diameter interpretative criteria (mm) |
MIC interpretative criteria (μg/ml) |
||||||||||
| S | I | R | S | I | R | S | I | R | S | I | R | ||||
| Enterobacteriaceae | Polymyxin B | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Colistin | — | — | — | — | — | — | — | — | — | — | — | ≤2 | — | >2 | |
| Pseudomonas spp. | Polymyxin B | — | — | — | — | ≤2 | 4 | ≥8 | — | — | — | — | — | — | — |
| Colistin | — | — | — | — | ≤2 | — | ≥4 | — | — | — | — | ≤2 | — | >2 | |
| Acinetobacter spp. | Polymyxin B | — | — | — | — | ≤2 | — | ≥4 | — | — | — | — | — | — | — |
| Colistin | — | — | — | — | ≤2 | — | ≥4 | — | — | — | — | ≤2 | — | >2 | |
S, susceptible; I, intermediate; R, resistant; —, not determined.
CLSI has recently established the MIC epidemiological cutoff value (ECV) for colistin applicable to five members of the Enterobacteriaceae, including E. coli, K. pneumoniae, K. aerogenes (formerly Enterobacter aerogenes), E. cloacae, and Raoultella ornithinolytica. ECVs are generally the MIC or zone diameter value that divides bacterial populations into those with and those without phenotypically detectable resistance (non-wild-type [NWT] and wild-type [WT], respectively). It is noteworthy that ECVs must not be used as clinical breakpoints for susceptible, intermediate, or resistant. If the colistin MIC for the above-mentioned members of the Enterobacteriaceae is at or below the ECV (≤2 μg/ml), it can be assumed that the isolate is a WT strain. If the colistin MIC is ≥4 μg/ml, the isolate should be retested, and if the result is confirmed, the isolate can be considered NWT.
Both CLSI and EUCAST recommend use of the quality control (QC) organisms E. coli ATCC 25922 (MIC range, 0.25 to 2 μg/ml) and P. aeruginosa ATCC 27853 (MIC range, 0.5 to 4 μg/ml) for polymyxin susceptibility testing to standardize the procedures (82, 85). Although there are many conflicting results between broth microdilution (BMD) and the gradient diffusion test, e.g., Etest, at MICs of ≥2 μg/ml, the suggested MICs of the QC strains are lower. In addition, EUCAST has currently advised laboratories to include colistin-resistant E. coli NCTC 13846 (mcr-1 positive) as a resistant QC strain. The recommended MIC target value of colistin for this strain is 4 μg/ml and should only occasionally be 2 μg/ml or 8 μg/ml (85). So, it is important that laboratories be aware of the possibility of the presence of mcr in isolates with colistin MICs of ≥2 μg/ml, indicating that further confirmation testing of isolates may be warranted.
Susceptibility Testing Methods for Polymyxins.
Despite the increasing use of polymyxins in clinical practices, the appropriate method for determining polymyxin susceptibility in routine clinical laboratories remains unclear (86). So far, several studies evaluating the performance of methods for polymyxin susceptibility testing have obtained inconsistent results; hence, there is a great challenge for the scientific community to develop a rapid and reliable method to determine the susceptibility of isolates to polymyxins (87).
The disk diffusion (DD) test, or Kirby-Bauer procedure, is a simple and inexpensive method for screening a large number of isolates to determine the antimicrobial susceptibility in many clinical laboratories (88). However, the poor and slow diffusion of polymyxins through agar is associated with small zones of growth inhibition and significant assay variation, negating use of this method for susceptibility testing (89, 90). In fact, the predictive accuracy of the DD test is unacceptable, and consequently, no reliable correlation of zone diameters and MICs has been found in many previous studies (89–92). The difficulty in differentiating the inhibition zone diameters has been illustrated by the study of Jerke et al. (93), where a resistant P. aeruginosa isolate with a zone diameter of 10 mm was compared to a susceptible isolate with a zone diameter of 11 mm; both isolates were categorized as susceptible with BMD MICs of ≤0.25 μg/ml. Additionally, studies have reported very major errors (VMEs), indicating false susceptibility, obtained from the DD test at rates ranging from 0.7% (92) to 3.5% (91) for colistin and from 1.2% (94) to 5% (95) for polymyxin B and with a low level of reproducibility (90) in comparison with the results of the BMD reference method. These findings explain why no zone diameter breakpoints are currently provided by CLSI and EUCAST for polymyxins (82, 83) and, thus, why the joint CLSI-EUCAST Polymyxin Breakpoints Working Group does not recommended use of the DD method for susceptibility testing of polymyxins (84).
The agar dilution (AD) susceptibility method relies on various concentrations of polymyxins in Mueller-Hinton agar (MHA) plates (2-fold dilution series). A bacterial inoculum corresponding to a 0.5 McFarland standard concentration is added as a spot to a plate with 104 CFU per spot. After incubation of the plates for 18 to 24 h at 35 ± 2°C, the bacterial growth in the inoculated spots is examined (88). This technique allows replicate spots of one bacterial strain or spots of different strains to be tested so that the MIC of the antibiotic against multiple strains can be tested on the same plate. However, AD is very laborious and is rarely used by many clinical laboratories.
Although AD may theoretically avoid the adsorption of polymyxins to the plates, no studies have measured the antibiotic concentration in agar dilution plates. One study (96) evaluating colistin stability in agar found that there were no differences in the MICs of two ATCC control strains obtained from 1-week-old colistin agar plates and from freshly used plates. In particular, the MICs obtained from all plates were 0.25 μg/ml and 2 μg/ml for E. coli ATCC 25922 and P. aeruginosa ATCC 27853, respectively. The results for colistin-resistant K. pneumoniae isolates on the plates that been stored for 1 week were also uniform and showed MICs of 128 μg/ml. These findings suggest that AD is a reliable and reproducible method to determine the colistin MIC, even when colistin-containing agar plates are stored for 1 week at 4°C. Additionally, the main findings of the study by Moskowitz et al. (97) indicated that AD is the most sensitive method to detect polymyxin resistance in clinical P. aeruginosa isolates. Other susceptibility testing methods, including BMD, DD, and Etest strips, exhibited high rates of significant false susceptibility for both P. aeruginosa and S. maltophilia clinical isolates compared with those achieved with AD.
Previous studies have also found a good accordance between the results of AD and BMD (90, 92, 98). In the study of 61 carbapenem-nonsusceptible clinical isolates, including 41 K. pneumoniae and 20 A. baumannii isolates (99), AD showed colistin susceptibility rates similar to those of BMD (4.9% versus 3.3%), with an acceptable categorical agreement (CA) (91.8%) being observed for both pathogens. In addition, relatively limited errors, including 3.3% VMEs and 4.9% major errors (MEs), were found by AD. Similarly, Behera et al. (92) found a concordance of 97% between AD and BMD for testing of the susceptibility to polymyxin B in 257 MDR Gram-negative bacteria, with 0.7% VMEs and 2.4% minor errors (mEs) for AD. However, trends toward higher MICs resulting from AD than from BMD have been described previously (99, 100). This can be due to the different concentrations of cations present in MHA, which is not suggested for use by CLSI guidance. Altogether, the joint CLSI-EUCAST Polymyxin Breakpoints Working Group recently did not recommend the use of AD for susceptibility testing of colistin (84).
BMD is widely employed as the reference test method for MIC susceptibility testing of all antimicrobials (88). As the CLSI-recommended method, BMD is performed with cation-adjusted Mueller-Hinton broth (CA-MHB), a range of doubling dilutions of antibiotic, and a final inoculum size of 5 × 105 CFU/ml in round or conical wells of a sterile plastic microdilution plate or tray. The MIC value is determined to be the lowest concentration of antibiotic that stops visible bacterial growth after 18 to 24 h of incubation at 35 ± 2°C (88). In particular, each well should contain 0.1 ml of broth, and the inoculum volume should be 0.01 ml. To prepare the inoculum, a 0.5 McFarland suspension should be diluted 1:20 to yield 5 × 106 CFU/ml. When 0.01 ml of this suspension is inoculated into the broth, the final test concentration of bacteria is approximately 5 × 105 CFU/ml (88).
Recently, the joint CLSI-EUCAST Subcommittee on Polymyxin Susceptibility Testing and Breakpoints recommended the ISO-20776 standard BMD method (with sulfate salts of colistin in plain polystyrene trays without additives, like polysorbate 80) as the ideal method to evaluate susceptibility to polymyxins (84). It has been claimed that BMD is time-consuming and requires expert staff for manual preparation of the antibiotic solutions for a routine clinical laboratory. Additionally, some technical issues for testing have been reported using this assay (101, 102). Polymyxins readily adhere to the plastic trays, leading to decreased antibiotic concentrations actually being present in the MHB dispensed in the wells (98, 103). The addition of a surfactant, such as polysorbate 80 (P-80), limits antibiotic adhesion to the BMD panels (104). Nonetheless, CLSI or EUCAST guidelines do not currently recommend the use of P-80 for colistin or polymyxin B susceptibility testing by BMD (84). As will be discussed further, it has been demonstrated that P-80 displays a synergistic effect with polymyxins (105) and has antibacterial activity of its own (106).
Another technical issue with polymyxin susceptibility testing using BMD is the presence of a “skipped well,” which makes it difficult to determine the true MIC. The phenomenon of a skipped well, which refers to the well without growth, despite the occurrence of growth in wells with higher concentrations, has been reported for Enterobacter species (107), P. aeruginosa (108), and A. baumannii (109). As presented in a study of Enterobacter species by Landman et al. (107), isolates with a multiple-skipped-well phenotype are associated with uninterpretable MIC results using the BMD assay. According to CLSI guidance (88), a single skipped well is acceptable and the well with the highest MIC value should be read. Isolates exhibiting more than one skipped well should be considered to have an uninterpretable polymyxin MIC; therefore, interpretation of susceptibility test results should be done with great care until the clinical relevance of this particular phenotype is defined. The phenomenon of a skipped well may represent heteroresistance and, in turn, might result from mutations affecting the response regulator PhoP (57, 107). This is supported by an experiment conducted by Jayol et al., where a colistin-resistant population of K. pneumoniae isolates that had a MIC of 128 μg/ml and that harbored the amino acid change Asp191Tyr in PhoP was found to coexist with a colistin-susceptible subpopulation (MIC = 0.12 μg/ml) (57). Furthermore, an increase in the expression of the phoQ, arnB, and pmrAB genes was demonstrated in the skipped-well isolates of P. aeruginosa, resulting in LPS modifications and a reduction of polymyxin-binding sites (108).
Nevertheless, BMD is currently the gold standard method for determination of the MICs of antimicrobial agents, including polymyxins, due to its reproducibility, reliability, and possibility of automation. A number of semiautomated devices for colistin BMD testing have been suggested for routine clinical practices (99, 110), although some problems have been reported with colistin susceptibility testing on these systems (111, 112). A study evaluating five commercially BMD products (Sensititre, Micronaut-S, Micronaut MIC-Strip, SensiTest, and UMIC) in comparison with the BMD reference method reported the rate of essential agreement (EA) to range from 82% to 99%, the rate of CA to range from 89% to 95%, the major error (ME) rate to range from 3% to 7%, and the VME range to be up to 3% (110). Another study by Chew et al. indicated that commercial and automated BMD systems may obtain results relatively comparable to those of the BMD reference method (79). Conversely, Vourli et al. (112) assessed the performance of two automated systems (Phoenix 100 and Vitek 2) for colistin susceptibility testing among carbapenem-resistant A. baumannii isolates. High rates of false susceptibility or VMEs were observed for both the Phoenix 100 (41.4%) and Vitek 2 (37.9%) systems. VMEs were much more frequent for isolates that showed MICs of 2 μg/ml than for isolates that showed MICs of ≤1 μg/ml, indicating that MIC results by automated systems within the susceptible range should be validated by a BMD assay. Until now, the U.S. Food and Drug Administration (FDA) has not approved the commercial automated assays for use for routine polymyxin susceptibility testing at clinical laboratories. This is because there are no FDA-recognized breakpoints for Gram-negative bacilli, so these methods are recommended only for research use.
Broth macrodilution (BMAD) is a method for the determination of MICs that is identical to BMD in terms of the growth medium (CA-MHB), the bacterial suspension used as the inoculum, the preparation of serial 2-fold dilutions of antimicrobial agent, the incubation conditions, and the reading of the plate; the methods differ by the volume of growth medium used and the use of sterile 13- by 100-mm test tubes instead of plastic trays (88). To prepare an adjusted inoculum, the 0.5 McFarland standard suspension is diluted 1:150, resulting in a tube containing approximately 5 × 106 CFU/ml. Addition of 1 ml of the standardized inoculum to each tube containing 1 ml of antimicrobial agent in the dilution series brings the final inoculum to 5 × 105 CFU/ml.
As the reference method, BMAD has been used to evaluate the accuracy of a commercially available BMD tray for detecting colistin and polymyxin B resistance in carbapenem-resistant K. pneumoniae isolates (113). Hindler and Humphries (98) evaluated the colistin BMAD method against the BMD method with P. aeruginosa (n = 60), K. pneumoniae (n = 20), and A. baumannii (n = 27) clinical isolates. They found no false-susceptible results and the highest essential agreement (EA) of 83% compared with the results of the other methods tested.
The gradient diffusion test, such as the Etest (bioMérieux) or M.I.C.Evaluator (Oxoid), is a variation on MIC determination. This test uses an inert and a nonporous plastic strip impregnated with the increasing concentrations of antibiotic (ranging from 0.016 to 256 μg/ml and 0.064 to 1,024 μg/ml for colistin and polymyxin B, respectively) to determine the MICs of different organisms. This method is performed by applying the strip to the surface of an MHA plate inoculated with a 0.5 McFarland standard. After incubation for 18 to 24 h at 35 ± 2°C, a symmetrical inhibition ellipse centered along the strip is formed, and the MIC value is defined from where the ellipse edge intersects the test strip (114, 115).
A variety of previous studies demonstrated a good correlation between the Etest method and the reference BMD technique (90–92, 116). Recently, a study with 202 carbapenem-resistant A. baumannii isolates found a high CA of 99.5% between the colistin Etest and BMD with P-80, whereas the CA between Etest and AD was 90.6% (117). In contrast, different studies have reported high error rates of the Etest method for the determination of polymyxin susceptibility (97–99, 107). Chew et al. (79) compared Etest for polymyxin susceptibility testing to BMD with 66 carbapenem-resistant isolates of the Enterobacteriaceae. EAs of 75% and 48.7% between these two methods were reported for colistin and polymyxin B, respectively. VME rates of 12% for colistin and 26.1% for polymyxin B were also found. In addition, the Etest method may misestimate the MIC values of polymyxins for test bacteria. Hindler and Humphries (98) revealed that Etest MICs compared to those obtained by BMD with P-80 were significantly lower among the colistin-resistant isolates (MICs ≥ 4 μg/ml), whereas MICs for susceptible isolates were raised (MICs < 4 μg/ml). Similar to what has been known for disk diffusion, this problem could be due to the poor diffusion of polymyxins into agar. Therefore, even though the gradient diffusion test is simple to perform, it is rather expensive and fails to reliably detect polymyxin-resistant strains.
More recently, a simplified approach, namely, colistin broth disk elution (CBDE), for colistin susceptibility determination has been described by Simner et al. (118). This method uses four, 10-ml CA-MHB tubes per isolate to which 0, 1, 2, and 4 colistin 10-μg disks are added, generating a final concentration of 0 (growth control), 1, 2, and 4 μg/ml in the tubes, respectively. After 30 min of incubation at room temperature, each tube is inoculated with a 50-μl aliquot of a 0.5 McFarland standard suspension for a final inoculum of 7.5 × 105 CFU/ml. MIC values are determined following 16 to 20 h of incubation at 35 ± 2°C. Data from the study of Simner and coworkers (118) revealed that CBDE yielded a CA and an EA of 98% and 99%, respectively, compared to the results of colistin BMD for the 172 Gram-negative bacilli tested. In addition, a CA and an EA of 100% each was achieved by CBDE compared to the results of BMAD. The overall VME rate was 8% because 3 of the E. coli mcr-1 isolates showed a MIC of 2 μg/ml by CBDE and a MIC of 4 μg/ml by BMD (118). These findings recommend that colistin MICs of 2 μg/ml by CBDE be confirmed by the reference BMD method and that isolates with MICs of ≥2 μg/ml be assessed for mcr genes.
Multicomponent Composition of Polymyxins.
Earlier works have suggested considerable variability in the chemical compositions of colistin and polymyxin B from the different manufacturers (119, 120). The heterogeneity in polymyxin formulations could affect their pharmacodynamic activity and, therefore, impact preclinical studies, including time-kill experiments and MIC testing, as well as clinical treatment. In a study by Diep et al. (121), there was a significant difference in the in vitro activity between several polymyxin B products, including those from Sigma-Aldrich, AK Scientific, USP, and MP Biomedicals. For instance, concentrations of 1 and 4 μg/ml of all products showed the same early killing activity against A. baumannii ATCC 19606. However, at 16 μg/ml, the Sigma-Aldrich polymyxin B exhibited more rapid bacterial killing at 2 h and 4 h than did the other products. Furthermore, the MP Biomedicals and USP polymyxin B products at 1 and 4 µg/ml resulted in the more rapid killing of K. pneumoniae ATCC BAA1705 by 2 h than the AK Scientific and Sigma-Aldrich products. At a 16-μg/ml concentration of polymyxin B, the MP Biomedicals product demonstrated more rapid killing at 2 h than the AK Scientific product.
Another important issue is the use of the inactive prodrug colistimethate sulfate (CMS) instead of colistin sulfate for MIC measurement. Due to doubts over whether CMS has antimicrobial activity itself, in vitro studies determining MICs have been performed by using colistin sulfate and CMS (23, 120). In a study investigating the in vitro pharmacodynamic properties of colistin and colistin methanesulfonate against clinical isolates of P. aeruginosa, Li et al. (120) found that the MICs of colistin for the susceptible strains ranged from 1 to 4 μg/ml, while the MICs of colistin methanesulfonate were significantly higher and ranged from 4 to 16 μg/ml. In addition, Bergen et al. (23) demonstrated that the concentration of colistin generated in 240 min after spiking of the samples with CMS at 32 μg/ml had the same killing effects as the colistin generated after spiking of the samples with CMS at 6 μg/ml by 15 min. An in vitro study (24) showed the hydrolysis of CMS to colistin in water, isotonic phosphate buffer, and human plasma within 24 to 48 h at 37°C. No formation of colistin was also observed when a 1,024-μg/ml concentration of CMS was stored for 30 min in MHB at 37°C. From these evidences, it is inappropriate to use CMS for MIC measurement, as antibacterial activity is due to the formation of colistin and not to the CMS in its own right.
Cationic Properties of Polymyxins.
It is well-known that the cationic nature of polymyxins causes them to adhere to a variety of laboratory plastic devices, including microtiter plates, leading to different interlaboratory results for the MIC by the BMD method (101, 102). The rate of drug binding depends on various factors, such as the nature of the plastic products and the coating used on the plates (81, 93, 101, 122). For example, tissue culture-coated microplates resulted in an overall 5.3-fold increase in the MIC values of colistin for all tested isolates (P. aeruginosa, Enterobacteriaceae, and Acinetobacter spp.), maybe due to the reduced free drug concentration within the microwells (101). Plastics can also be manufactured as microtubes with the ability to minimize any interactions between the analytes and the tubes, and these are often marketed as low-protein/DNA-binding microtubes. In recent experiments with low-protein-binding polypropylene microtubes (122), the initial concentrations of colistin were higher than those in other materials (a standard polypropylene tube, polystyrene tube, glass tube, and standard polystyrene microplate), at between 63% and 99%. These type of microtubes showed the least adsorption, followed by standard polypropylene and glass. Similar results have also been obtained from other studies, demonstrating that BMD with glass-coated plates gives more reliable MIC results for polymyxins than BMD with polystyrene plates (96, 123). Furthermore, Karvanen et al. (103) previously showed that colistin adsorption was proportionally higher at a lower concentration of 0.125 μg/ml after 24 h at 35°C, with the estimated colistin concentration in MHB being 8%, 13%, and 25% in polystyrene, polypropylene, and glass-coated wells, respectively. However, at an initial concentration of 4 μg/ml, 75%, 62%, and 62% of the colistin remained following 24 h in polystyrene, polypropylene, and glass, respectively. These findings indicate that polymyxins are extensively lost under normal experimental conditions in a concentration- and material-dependent manner; thus, it is important to carefully monitor drug concentrations during the experiment course in order to gain more accurate results for the antibacterial effect of antibiotic.
Effect of Polysorbate 80.
Polysorbate 80 (P-80 or Tween 80) is a nonionic surfactant and emulsifier often used in foods and cosmetics. Structurally, this surfactant contains both a hydrophilic polyether group and a lipophilic oleic acid chain (93). It is also used in bacterial broth cultures to prevent foam formation and as an excipient in many medications and vaccines against influenza to stabilize aqueous formulations (124). The antimicrobial properties of microemulsions containing surfactant molecules, in particular, P-80, have been reported in some studies published in the literature, where such microemulsions caused a complete loss of viability of Staphylococcus aureus and E. coli (125) and effectively decreased the viability of an established biofilm population of P. aeruginosa (124). Furthermore, the synergistic effect of P-80 with various chemical agents, such as benzalkonium chloride, chlorhexidine diacetate, and polymyxin B, which leads to reduced resistance to these agents, has been previously described in P. aeruginosa (105, 126). P-80 also has a mild antimicrobial activity, especially when it is combined with other antimicrobial agents; increases cell permeability; and lyses spheroplasts (106). It is feasible that the primary damage to the outer membrane caused by polymyxins allows the surfactant to enter the cell and promote cell lysis. Therefore, polymyxin-resistant bacteria would not be affected by P-80, as their cell membrane remains intact, giving higher MICs (127). This can be an explanation for the findings that P-80 mainly affects isolates showing MICs of ≤1 or ≤2 μg/ml (98, 104).
A CLSI guideline has recommended the use of P-80 as a dispersing agent at a final concentration of 0.002% for the testing of some lipoglycopeptides, including dalbavancin, oritavancin, and telavancin, to prevent the binding of drug to plastics (82, 88). Studies have also demonstrated that the addition of P-80 either to the bacterial inoculum or directly to MHB minimizes polymyxin adhesion to microplates, improving MIC susceptibility testing results for both polymyxin B and colistin (98, 128, 129). In a study with 247 organisms, including E. coli, K. pneumoniae, P. aeruginosa, and Acinetobacter isolates, Sader et al. (104) found a significant downward shift (4- to 8-fold) in the MICs of polymyxins measured by BMD with the addition of a 0.002% final concentration of P-80. The decreases were also the most appreciable for susceptible organisms having MICs of ≤2 μg/ml compared with those for nonsusceptible strains with MICs of >2 μg/ml, indicating that the reduction in the MIC in the presence of surfactant does not change remarkably the susceptibility category when the current CLSI breakpoints are applied to polymyxins. Regardless, the CLSI subcommittee decided in January 2014 to adopt polymyxin (colistin) BMD testing without the addition of P-80 (87). The outcomes of this decision will hinge mostly on the breakpoints adopted for the polymyxins. If the present susceptibility breakpoints of ≤2 or ≥4 μg/ml are retained, the MIC interpretation is unlikely to be compromised. However, the ability of BMD to detect susceptible isolates without using P-80 may be weakened, if a susceptible breakpoint of ≤1 μg/ml is chosen. Because the MIC results obtained by BMAD in glass tubes, which have the lowest colistin adsorption, are comparable to those obtained by BMD with P-80, determination of polymyxin MICs in glass-coated plates could address this concern (98).
Altogether, since the potential antimicrobial activity of Tweens alone was not exactly explored, the use of P-80 to evaluate the susceptibility of Gram-positive pathogens (vancomycin- and methicillin-resistant staphylococci, vancomycin-resistant enterococci, and penicillin-resistant streptococci) to some lipoglycopeptides, including dalbavancin, oritavancin, and telavancin, is recommended to be continued (82, 130, 131).
The Heteroresistance Phenomenon.
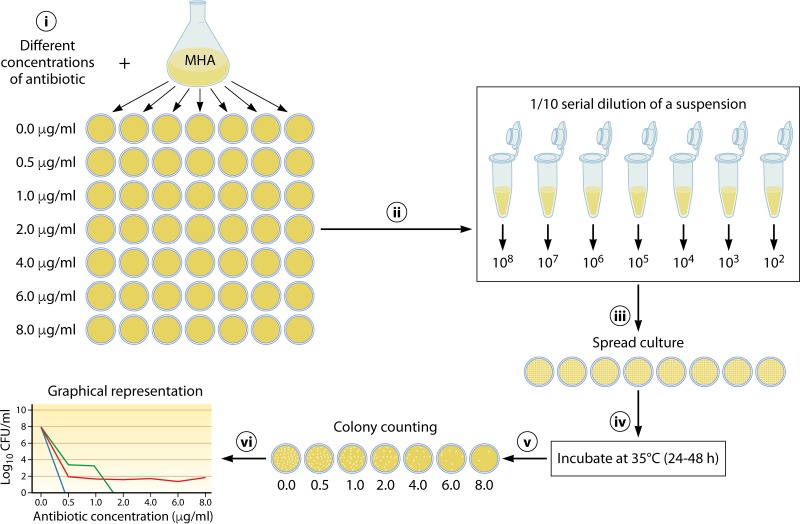
The phenomenon of heteroresistance could pose a new challenge to antimicrobial therapy, especially from the point of view that it is not detected by the traditional susceptibility testing methods and may also be misinterpreted on susceptibility categorization (61). While population analysis profiling (PAP) is considered the gold standard method broadly used to characterize heteroresistant bacteria (60, 62, 107, 132–134), other susceptibility assays, such as DD, BMD, the gradient diffusion test, and commercial automated systems, have also been employed (59, 90, 109, 135, 136). PAP analysis is usually performed by inoculation of a bacterial population onto MHA plates containing gradient concentrations of antibiotic and quantifying the bacterial growth at each of these concentrations. To facilitate counting of the number of CFU in each plate after 48 h of incubation at 35°C, serial saline dilutions of an overnight culture are spread on MHA plates (Fig. 4) (62, 133). The detection of heteroresistance by the PAP method for clinical laboratories is difficult and time-consuming. Furthermore, there is no standard guideline to perform PAP; in particular, there is no guideline for the selection of antibiotic concentration increments, such as 1- to 2-μg/ml steps (55, 60, 62, 137) and even steps as low as 0.1 μg/ml (138), or the starting concentration of the bacterial inoculum (0.5 McFarland and even higher) (55, 58, 133, 139, 140), leading to categorization errors (137). DD and gradient diffusion strip tests have been used for determining heteroresistance, as recommended for traditional in vitro susceptibility testing (57–59, 133, 135). A criterion for heteroresistance is the growth of colonies within the inhibition zone of disks and/or gradient diffusion strips (135). In a study by Lo-Ten-Foe et al. (90), Etest and DD were found to be methods advantageous for the detection of colistin-heteroresistant E. cloacae and A. baumannii isolates. Jayol et al. (57) showed that Etest strips can sufficiently identify heteroresistance to colistin in K. pneumoniae isolates with PhoP-PhoQ alterations. On the other hand, Gravey et al. (139) evaluated the performance of various techniques for detecting colistin-heteroresistant E. cloacae clinical isolates. By comparison with PAP, DD and Etest showed low sensitivities of 22.9% and 54.2%, respectively, although the specificity for both methods was 100% (139). The low sensitivity of DD and Etest to detect heteroresistant strains might be due to the low bacterial load used in these methods, in that the load is improper for detecting the resistant subpopulations present at a low frequency. In addition, automated systems did not exhibit appropriate performance with bacteria with resistant subpopulations. Supporting this claim is the finding of a study by Lo-Ten-Foe et al. (90), where the Vitek 2 susceptibility test failed to detect colistin-heteroresistant E. cloacae isolates. The authors considered it necessary to use an alternative testing method with the capacity to detect resistant subpopulations for isolates in which occasional heteroresistance has been described.
FIG 4.
Population analysis profile (PAP) protocol to detect the heteroresistance phenomenon in bacteria. PAP is usually performed by inoculation of a bacterial population onto MHA plates containing increasing concentrations of antibiotic, quantifying the bacterial growth at each antibiotic concentration, and, finally, graphical analysis of the results.
The Impact of Medium.
The cation composition of Mueller-Hinton (MH) medium has been shown to vary depending on the commercial brand (141). According to CLSI, a calcium concentration ranging from 20 to 25 mg/liter and a magnesium concentration ranging from 10 to 12.5 mg/liter in MH medium ensures reliable antimicrobial susceptibility results (88). A study by Girardello et al. (142) demonstrated that the calcium and magnesium concentrations measured for four MHB brands (Oxoid, Difco, Merck, HiMedia) were far below the recommendations of the CLSI guidance. None of the MHA commercial brands tested showed the correct concentrations of calcium and magnesium recommended by the CLSI. Although an EA of 100% was found for the Difco medium used, the EA rates between BMD and Etest for determination of the polymyxin B MIC was reported to be 80% for the Merck and Oxoid MHAs and 90% for the HiMedia MHA. In addition, the Merck MHA produced zones of inhibition larger than those produced by the other MHA brands tested. A poor EA of 46%, 64%, and 68% between the Etest and BMD with P-80 was reported by Hindler and Humphries (98) for the BBL, Hardy, and Remel MHA brands, respectively. However, CA was 78% for the BBL MHA, 78% for the Hardy MHA, and 84% for the Remel MHA. Such discrepancies in antimicrobial susceptibility testing results may be due to the distinct cation concentrations in the MHA or MHB brands tested (141, 143, 144). This is why the CLSI recommends the use of cation-adjusted Mueller-Hinton (CA-MH) medium or supplementation of the culture medium with cations for antimicrobial susceptibility testing purposes (88). Moreover, a study by Matzneller et al. (145) revealed that colistin MIC values against P. aeruginosa and A. baumannii increased significantly in a cation-dependent manner. In contrast, colistin activity against E. coli showed a linear increase with ascending cation concentrations. These findings indicate that the antibacterial activity of colistin is cation dependent and might be misestimated. A medium, namely, Iso-Sensitest agar (ISA), was introduced by the British Society for Antimicrobial Chemotherapy (BSAC) to overcome problems associated with the traditional media used for antimicrobial susceptibility testing procedures (146, 147). This medium allows the growth of the vast majority of microorganisms without additional supplementation. For fastidious organisms, it can also be supplemented with 5% whole horse blood and 20 mg/liter beta-NAD (148). Furthermore, ISA medium has been shown to be better than other media for the detection of heteroresistance. Lo-Ten-Foe et al. (90) compared the AD and Etest methods for colistin on MHA and ISA media. Their study demonstrated that the Etest on MHA failed to detect resistant subpopulations of heteroresistant E. cloacae isolates, while these resistant subpopulations were detected on ISA by both methods, reflecting the higher sensitivity of ISA for the detection of resistant subpopulations. For these reasons, there is still no consensus on the use of cation-adjusted or non-cation-adjusted medium, and thus, a consensus is needed.
Detection of mcr-1-carrying isolates by phenotypic methods using CA-MH medium with BMD remains a challenge for routine microbiology laboratories, where many isolates exhibit a MIC of 2 μg/ml to colistin and are thus categorized as sensitive. More recently, Gwozdzinski et al. developed a medium with a novel formulation called calcium-enhanced Mueller-Hinton (CE-MH) medium containing 200 mg/liter Ca2+ ions with the capability of the simple and reliable determination of the colistin MIC in mcr-1-producing Enterobacteriaceae (149). In addition, this medium allowed the identification of colistin-resistant isolates that were incorrectly classified as susceptible by the reference BMD method in CA-MH. Further genomic analysis of these isolates revealed specific alterations in the pmrCAB operon, confirming the efficacy of CE-MH for colistin resistance testing compared to that of the reference medium, CA-MH (149).
The Impact of Bacterial Storage.
The loss of colistin resistance following ≤12 months of storage in glycerol-supplemented brucella broth at −70°C has been reported by Hindler and Humphries (98). They found that 5 out of 14 A. baumannii, K. pneumoniae, and P. aeruginosa isolates that were initially resistant to colistin became susceptible after frozen storage. In study by Barin et al. (55), the polymyxin B MICs of all 4 A. baumannii isolates tested growing at 64 μg/ml was reduced to 0.125 to 0.25 μg/ml after 60 days of storage at −80°C. These findings indicate that polymyxin resistance might be lost following long-term storage.
The Impact of Bacterial Subcultures.
The loss of resistance has been shown after the serial passaging of in vitro-selected colistin-resistant mutants without selective pressure (55, 62). Li et al. (62) demonstrated the loss of adaptive resistance to colistin in about 98% and 97% of colistin-resistant A. baumannii subpopulations, including ATCC 19606 and a clinical isolate, respectively, following 16 successive passages in colistin-free CA-MHB medium. Adaptive resistance to polymyxins, which is characterized by the induction of resistance in the presence of drug and reversal to the susceptible phenotype in its absence, has been identified in other Gram-negative bacilli (150, 151). Reversal to colistin susceptibility in P. aeruginosa has been observed in one study (150) by serial passage of strains with a phenotype of adaptive resistance to colistin in drug-free medium. Structural analysis revealed that the lipid A modification with l-Ara4N and palmitate observed in the evolved colistin-resistant strains did not appear in the revertant strains. Sequence analysis also demonstrated the lack of some genetic alterations within several TCSs of the colistin-susceptible revertants; all of those alterations were observed in colistin-resistant mutants. Reversions to the wild-type amino acid in the colistin-susceptible P. aeruginosa revertants under drug-free conditions might explain the restoration of colistin susceptibility (150). This acquisition and loss of polymyxin resistance might be one of the reasons for the variable results obtained from laboratory susceptibility testing, leading to the clinical ineffectiveness of the antibiotic.
CONCLUSIONS
The growing threat of infections caused by multiresistant Gram-negative bacilli, particularly in vulnerable patients, has currently forced many physicians to turn to polymyxins. This emphasizes the need for the effective evaluation of critical issues associated with testing for susceptibility to these compounds. Because of the unreliable results obtained from both DD and gradient diffusion susceptibility tests, BMD remains the only option for the in vitro assessment of polymyxin susceptibility. The adherence of polymyxins to the plastics used for BMD trays is an issue leading to significant variability in MIC results. Although addition of the surfactant P-80 alleviates antibiotic adsorption, the joint CLSI-EUCAST Polymyxin Breakpoints Working Group does not recommend that because of the synergistic activity of P-80 with polymyxins. Moreover, whether cation-adjusted or non-cation-adjusted medium should be used and whether BMD should be performed with or without P-80 have not yet been fully defined. The issue of brand-to-brand and batch-to-batch heterogeneity in polymyxin formulations is also a source of difference in MIC results. Resistance to polymyxins is increasingly being reported among MDR clinical isolates worldwide. A variety of LPS modifications, mainly mediated by phoP-phoQ and pmrA-pmrB TCSs, are strategies to protect bacteria from polymyxins. However, there is still a knowledge gap in a full understanding of polymyxin resistance, particularly in areas of heteroresistance and the conditions required for the activity of mcr genes. Further research is required to clarify (i) the optimal method for polymyxin susceptibility testing in routine diagnostic settings, (ii) the impact of heteroresistance on susceptibility testing results and, (iii) the correlation between polymyxin MICs and clinical outcomes to guide decision-making for the treatment of MDR Gram-negative bacterial infections.
ACKNOWLEDGMENTS
We appreciate the financial support of Golestan University of Medical Sciences, Gorgan, Iran.
REFERENCES
- 1.O’Neill J. 2016. Review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. http://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf.
- 2.Smith RD, Coast J. 2002. Antimicrobial resistance: a global response. Bull World Health Organ 80:126–133. [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal S, Kakati B, Khanduri S, Gupta S. 2017. Emergence of carbapenem resistant non-fermenting gram-negative bacilli isolated in an ICU of a tertiary care hospital. J Clin Diagn Res 11:DC04–DC07. doi: 10.7860/JCDR/2017/24023.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lari AR, Ardebili A, Hashemi A. 2018. AdeR-AdeS mutations & overexpression of the AdeABC efflux system in ciprofloxacin-resistant Acinetobacter baumannii clinical isolates. Indian J Med Res 147:413–421. doi: 10.4103/ijmr.IJMR_644_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razavi Nikoo H, Ardebili A, Mardaneh J. 2017. Systematic review of antimicrobial resistance of clinical Acinetobacter baumannii isolates in Iran: an update. Microb Drug Resist 23:744–756. doi: 10.1089/mdr.2016.0118. [DOI] [PubMed] [Google Scholar]
- 6.Paul M, Carmeli Y, Durante-Mangoni E, Mouton JW, Tacconelli E, Theuretzbacher U, Mussini C, Leibovici L. 2014. Combination therapy for carbapenem-resistant Gram-negative bacteria. J Antimicrob Chemother 69:2305–2309. doi: 10.1093/jac/dku168. [DOI] [PubMed] [Google Scholar]
- 7.Kwa A, Kasiakou SK, Tam VH, Falagas ME. 2007. Polymyxin B: similarities to and differences from colistin (polymyxin E). Expert Rev Anti Infect Ther 5:811–821. doi: 10.1586/14787210.5.5.811. [DOI] [PubMed] [Google Scholar]
- 8.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. 2013. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 10.Falagas ME, Kasiakou SK. 2006. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit Care 10:R27. doi: 10.1186/cc3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortwine JK, Kaye KS, Li J, Pogue JM. 2015. Colistin: understanding and applying recent pharmacokinetic advances. Pharmacotherapy 35:11–16. doi: 10.1002/phar.1484. [DOI] [PubMed] [Google Scholar]
- 12.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 13.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 15.Michalopoulos AS, Karatza DC. 2010. Multidrug-resistant Gram-negative infections: the use of colistin. Expert Rev Anti Infect Ther 8:1009–1017. doi: 10.1586/eri.10.88. [DOI] [PubMed] [Google Scholar]
- 16.Pournajaf A, Rajabnia R, Razavi S, Solgi S, Ardebili A, Yaghoubi S, Khodabandeh M, Yahyapour Y, Emadi B, Irajian G. 2018. Molecular characterization of carbapenem-resistant Acinetobacter baumannii isolated from pediatric burns patients in an Iranian hospital. Trop J Pharm Res 17:135–141. doi: 10.4314/tjpr.v17i1.19. [DOI] [Google Scholar]
- 17.Morrill HJ, Pogue JM, Kaye KS, LaPlante KL. 2015. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis 2:ofv050. doi: 10.1093/ofid/ofv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shariati A, Azimi T, Ardebili A, Chirani A, Bahramian A, Pormohammad A, Sadredinamin M, Erfanimanesh S, Bostanghadiri N, Shams S, Hashemi A. 2018. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microbes New Infect 21:75–80. doi: 10.1016/j.nmni.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2013. Antibiotic resistant threats in the United States, 2013. http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 20.He H, Li JC, Nation RL, Jacob J, Chen G, Lee HJ, Tsuji BT, Thompson PE, Roberts K, Velkov T, Li J. 2013. Pharmacokinetics of four different brands of colistimethate and formed colistin in rats. J Antimicrob Chemother 68:2311–2317. doi: 10.1093/jac/dkt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LF, Kaye D. 2009. Current use for old antibacterial agents: polymyxins, rifamycins, and aminoglycosides. Infect Dis Clin 23:1053–1075. doi: 10.1016/j.idc.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Barnett M, Bushby S, Wilkinson S. 1964. Sodium sulphomethyl derivatives of polymyxins. Br J Pharmacol Chemother 23:552–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergen PJ, Li J, Rayner CR, Nation RL. 2006. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:1953–1958. doi: 10.1128/AAC.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J, Milne RW, Nation RL, Turnidge JD, Coulthard K. 2003. Stability of colistin and colistin methanesulfonate in aqueous media and plasma as determined by high-performance liquid chromatography. Antimicrob Agents Chemother 47:1364–1370. doi: 10.1128/AAC.47.4.1364-1370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orwa J, Govaerts C, Gevers K, Roets E, Van Schepdael A, Hoogmartens J. 2002. Study of the stability of polymyxins B1, E1 and E2 in aqueous solution using liquid chromatography and mass spectrometry. J Pharm Biomed Anal 29:203–212. doi: 10.1016/S0731-7085(02)00016-X. [DOI] [PubMed] [Google Scholar]
- 26.Schupp J, Travis S, Price L, Shand R, Keim P. 1995. Rapid bacterial permeabilization reagent useful for enzyme assays. Biotechniques 19:18–20. [PubMed] [Google Scholar]
- 27.He J, Figueroa DA, Lim T-P, Chow DS, Tam VH. 2010. Stability of polymyxin B sulfate diluted in 0.9% sodium chloride injection and stored at 4 or 25° C. Am J Health Syst Pharm 67:1191–1194. doi: 10.2146/ajhp090472. [DOI] [PubMed] [Google Scholar]
- 28.Evans ME, Feola DJ, Rapp RP. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann Pharmacother 33:960–967. doi: 10.1345/aph.18426. [DOI] [PubMed] [Google Scholar]
- 29.Deris ZZ, Akter J, Sivanesan S, Roberts KD, Thompson PE, Nation RL, Li J, Velkov T. 2014. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J Antibiot 67:147–151. doi: 10.1038/ja.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain J-M. 2012. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 31.Gales AC, Jones R, Sader HS. 2006. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin Microbiol Infect 12:315–321. doi: 10.1111/j.1469-0691.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 32.Kuck N. 1976. In vitro and in vivo activities of minocycline and other antibiotics against Acinetobacter (Herellea-Mima). Antimicrob Agents Chemother 9:493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schülin T. 2002. In vitro activity of the aerosolized agents colistin and tobramycin and five intravenous agents against Pseudomonas aeruginosa isolated from cystic fibrosis patients in southwestern Germany. J Antimicrob Chemother 49:403–406. [DOI] [PubMed] [Google Scholar]
- 34.Bulman ZP, Zhao M, Satlin MJ, Chen L, Kreiswirth BN, Walsh TJ, Nation RL, Li J, Tsuji BT. 2018. Polymyxin B and fosfomycin thwart KPC-producing Klebsiella pneumoniae in the hollow fibre infection model. Int J Antimicrob Agents 52:114–118. doi: 10.1016/j.ijantimicag.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohlenberg A, Weitzel-Kage D, Van der Linden P, Sohr D, Vögeler S, Kola A, Halle E, Rüden H, Weist K. 2010. Outbreak of carbapenem-resistant Pseudomonas aeruginosa infection in a surgical intensive care unit. J Hosp Infect 74:350–357. doi: 10.1016/j.jhin.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Chen Y, Fang Y, Wang X, Chen Y, Qi Q, Huang F, Xiao X. 2015. Meta-analysis of colistin for the treatment of Acinetobacter baumannii infection. Sci Rep 5:17091. doi: 10.1038/srep17091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gales AC, Jones RN, Sader HS. 2011. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother 66:2070–2074. doi: 10.1093/jac/dkr239. [DOI] [PubMed] [Google Scholar]
- 38.Dogonchi AA, Ghaemi EA, Ardebili A, Yazdansetad S, Pournajaf A. 2018. Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: a potential threat to clinical therapeutics. Tzu Chi Med J 30:90–96. doi: 10.4103/tcmj.tcmj_101_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.European Centre for Disease Prevention and Control. 2015. Antimicrobial resistance surveillance in Europe 2014. Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). European Centre for Disease Prevention and Control, Stockholm, Sweden. [Google Scholar]
- 40.Sun S, Negrea A, Rhen M, Andersson DI. 2009. Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 53:2298–2305. doi: 10.1128/AAC.01016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrow K, Kwon DH. 2009. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:5150–5154. doi: 10.1128/AAC.00893-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayol A, Poirel L, Brink A, Villegas M-V, Yilmaz M, Nordmann P. 2014. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother 58:4762–4766. doi: 10.1128/AAC.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kline T, Trent M, Stead C, Lee M, Sousa M, Felise H, Nguyen HV, Miller SI. 2008. Synthesis of and evaluation of lipid A modification by 4-substituted 4-deoxy arabinose analogs as potential inhibitors of bacterial polymyxin resistance. Bioorg Med Chem Lett 18:1507–1510. doi: 10.1016/j.bmcl.2007.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunn JS. 2001. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res 7:57–62. [PubMed] [Google Scholar]
- 46.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H, Hsu FF, Turk J, Groisman EA. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol 186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nummila K, Kilpeläinen I, Zähringer U, Vaara M, Helander IM. 1995. Lipopolysaccharides of polymyxin B‐resistant mutants of Escherichia coli are extensively substituted by 2‐aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol Microbiol 16:271–278. doi: 10.1111/j.1365-2958.1995.tb02299.x. [DOI] [PubMed] [Google Scholar]
- 49.Kim S-H, Jia W, Parreira VR, Bishop RE, Gyles CL. 2006. Phosphoethanolamine substitution in the lipid A of Escherichia coli O157:H7 and its association with PmrC. Microbiology 152:657–666. doi: 10.1099/mic.0.28692-0. [DOI] [PubMed] [Google Scholar]
- 50.Helander IM, Kato Y, Kilpeläinen I, Kostiainen R, Lindner B, Nummila K, Sugiyama T, Yokochi T. 1996. Characterization of lipopolysaccharides of polymyxin‐resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem 237:272–278. doi: 10.1111/j.1432-1033.1996.0272n.x. [DOI] [PubMed] [Google Scholar]
- 51.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KRO, Doi Y, Ernst RK. 2013. Unique structural modifications are present in the LPS from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barin J, Martins AF, Heineck BL, Barth AL, Zavascki AP. 2013. Hetero- and adaptive resistance to polymyxin B in OXA-23-producing carbapenem-resistant Acinetobacter baumannii isolates. Ann Clin Microbiol Antimicrob 12:15. doi: 10.1186/1476-0711-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yau W, Owen RJ, Poudyal A, Bell JM, Turnidge JD, Yu HH, Nation RL, Li J. 2009. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J Infect 58:138–144. doi: 10.1016/j.jinf.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hjort K, Nicoloff H, Andersson DI. 2016. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol 102:274–289. doi: 10.1111/mmi.13459. [DOI] [PubMed] [Google Scholar]
- 59.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hermes DM, Pitt CP, Lutz L, Teixeira AB, Ribeiro VB, Netto B, Martins AF, Zavascki AP, Barth AL. 2013. Evaluation of heteroresistance to polymyxin B among carbapenem-susceptible and -resistant Pseudomonas aeruginosa. J Med Microbiol 62:1184–1189. doi: 10.1099/jmm.0.059220-0. [DOI] [PubMed] [Google Scholar]
- 61.Falagas M, Makris G, Dimopoulos G, Matthaiou D. 2008. Heteroresistance: a concern of increasing clinical significance? Clin Microbiol Infect 14:101–104. doi: 10.1111/j.1469-0691.2007.01912.x. [DOI] [PubMed] [Google Scholar]
- 62.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hernan RC, Karina B, Gabriela G, Marcela N, Carlos V, Angela F. 2009. Selection of colistin-resistant Acinetobacter baumannii isolates in postneurosurgical meningitis in an intensive care unit with high presence of heteroresistance to colistin. Diagn Microbiol Infect Dis 65:188–191. doi: 10.1016/j.diagmicrobio.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 64.Bardet L, Baron S, Leangapichart T, Okdah L, Diene SM, Rolain J-M. 2017. Deciphering heteroresistance to colistin in a Klebsiella pneumoniae isolate from Marseille, France. Antimicrob Agents Chemother 61:e00356-17. doi: 10.1128/AAC.00356-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. 2011. The insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3022–3024. doi: 10.1128/AAC.01732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 67.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 68.Gefen O, Balaban NQ. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol Rev 33:704–717. doi: 10.1111/j.1574-6976.2008.00156.x. [DOI] [PubMed] [Google Scholar]
- 69.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 70.Jeannot K, Bolard A, Plésiat P. 2017. Resistance to polymyxins in Gram-negative organisms. Int J Antimicrob Agents 49:526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 71.Skov RL, Monnet DL. 2016. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 21(9):pii=30155 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 72.Baron S, Hadjadj L, Rolain J-M, Olaitan AO. 2016. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 48:583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 73.Dalmolin TV, de Lima-Morales D, Barth AL. 2018. Plasmid-mediated colistin resistance: what do we know? J Infectiol 1:16–22. [Google Scholar]
- 74.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):pii=30280 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 75.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22(31):pii=30589 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma S, Sun C, Hulth A, Li J, Nilsson LE, Zhou Y, Börjesson S, Bi Z, Bi Z, Sun Q, Wang Y. 2018. Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J Antimicrob Chemother 73:1777–1780. doi: 10.1093/jac/dky110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Snesrud E, Maybank R, Kwak YI, Jones AR, Hinkle MK, McGann P. 2018. Chromosomally encoded mcr-5 in colistin-nonsusceptible Pseudomonas aeruginosa. Antimicrob Agents Chemother 62:e00679-18. doi: 10.1128/AAC.00679-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chew KL, La M-V, Lin RT, Teo JW. 2017. Colistin and polymyxin B susceptibility testing for carbapenem-resistant and mcr-positive Enterobacteriaceae: comparison of Sensititre, MicroScan, Vitek 2, and Etest with broth microdilution. J Clin Microbiol 55:2609–2616. doi: 10.1128/JCM.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. 2017. Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in south and southeast Asia. Clin Microbiol Rev 30:1–22. doi: 10.1128/CMR.00042-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing. CLSI document M100, 28th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 83.European Committee on Antimicrobial Susceptibility Testing. 2018. Breakpoint tables for interpretation of MICs and zone diameters, version 8.1 http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf.
- 84.CLSI-EUCAST Polymyxin Breakpoints Working Group. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 85.European Committee on Antimicrobial Susceptibility Testing. 2018. Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST, version 8.0 https://www.ipna.csic.es/sites/default/files/users/user282/EUCAST%202018.pdf.
- 86.Vasoo S. 2017. Susceptibility testing for the polymyxins: two steps back, three steps forward? J Clin Microbiol 55:2573–2582. doi: 10.1128/JCM.00888-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Humphries RM. 2015. Susceptibility testing of the polymyxins: where are we now? Pharmacotherapy 35:22–27. doi: 10.1002/phar.1505. [DOI] [PubMed] [Google Scholar]
- 88.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 10th ed M7-A10 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 89.An TY, Ng LSY. 2006. Comparison of three standardized disc susceptibility testing methods for colistin. J Antimicrob Chemother 58:864–867. doi: 10.1093/jac/dkl330. [DOI] [PubMed] [Google Scholar]
- 90.Lo-Ten-Foe JR, de Smet AMG, Diederen BM, Kluytmans JA, van Keulen PH. 2007. Comparative evaluation of the VITEK 2, disk diffusion, Etest, broth microdilution, and agar dilution susceptibility testing methods for colistin in clinical isolates, including heteroresistant Enterobacter cloacae and Acinetobacter baumannii strains. Antimicrob Agents Chemother 51:3726–3730. doi: 10.1128/AAC.01406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maalej S, Meziou M, Rhimi F, Hammami A. 2011. Comparison of disc diffusion, Etest and agar dilution for susceptibility testing of colistin against Enterobacteriaceae. Lett Appl Microbiol 53:546–551. doi: 10.1111/j.1472-765X.2011.03145.x. [DOI] [PubMed] [Google Scholar]
- 92.Behera B, Mathur P, Das A, Kapil A, Gupta B, Bhoi S, Farooque K, Sharma V, Misra MC. 2010. Evaluation of susceptibility testing methods for polymyxin. Int J Infect Dis 14:e596. doi: 10.1016/j.ijid.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 93.Jerke KH, Lee MJ, Humphries RM. 2016. Polymyxin susceptibility testing: a cold case reopened. Clin Microbiol Newsl 38:69–77. doi: 10.1016/j.clinmicnews.2016.04.003. [DOI] [Google Scholar]
- 94.van der Heijden IM, Levin AS, De Pedri EH, Fung L, Rossi F, Duboc G, Barone AA, Costa SF. 2007. Comparison of disc diffusion, Etest and broth microdilution for testing susceptibility of carbapenem-resistant P. aeruginosa to polymyxins. Ann Clin Microbiol Antimicrob 15:6–8. doi: 10.1186/1476-0711-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gales AC, Reis AO, Jones RN. 2001. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol 39:183–190. doi: 10.1128/JCM.39.1.183-190.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turlej-Rogacka A, Xavier BB, Janssens L, Lammens C, Zarkotou O, Pournaras S, Goossens H, Malhotra-Kumar S. 2018. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis 37:345–353. doi: 10.1007/s10096-017-3140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moskowitz SM, Garber E, Chen Y, Clock SA, Tabibi S, Miller AK, Doctor M, Saiman L. 2010. Colistin susceptibility testing: evaluation of reliability for cystic fibrosis isolates of Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Antimicrob Chemother 65:1416–1423. doi: 10.1093/jac/dkq131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug resistant Gram-negative bacilli. J Clin Microbiol 51:1678–1684. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dafopoulou K, Zarkotou O, Dimitroulia E, Hadjichristodoulou C, Gennimata V, Pournaras S, Tsakris A. 2015. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 59:4625–4630. doi: 10.1128/AAC.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hogardt M, Schmoldt S, Gotzfried M, Adler K, Heesemann J. 2004. Pitfalls of polymyxin antimicrobial susceptibility testing of Pseudomonas aeruginosa isolated from cystic fibrosis patients. J Antimicrob Chemother 54:1057–1061. doi: 10.1093/jac/dkh470. [DOI] [PubMed] [Google Scholar]
- 101.Albur M, Noel A, Bowker K, MacGowan A. 2014. Colistin susceptibility testing: time for a review. J Antimicrob Chemother 69:1432–1434. doi: 10.1093/jac/dkt503. [DOI] [PubMed] [Google Scholar]
- 102.Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, Tsuji BT, Turnidge JD. 2015. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 103.Karvanen MC, Mohamad A, Lagerback P. 2011. Colistin is extensively lost during normal experimental conditions, abstr D-690, p 160. Abstr 51st Intersci Conf Antimicrob Agents Chemother. American Society for Microbiology, Washington, DC. [Google Scholar]
- 104.Sader HS, Rhomberg PR, Flamm RK, Jones RN. 2012. Use of a surfactant (polysorbate 80) to improve MIC susceptibility testing results for polymyxin B and colistin. Diagn Microbiol Infect Dis 74:412–414. doi: 10.1016/j.diagmicrobio.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 105.Brown MR, Winsley BE. 1971. Synergism between polymyxin and polysorbate 80 against Pseudomonas aeruginosa. J Gen Microbiol 68:367–373. doi: 10.1099/00221287-68-3-367. [DOI] [PubMed] [Google Scholar]
- 106.Brown MR, Geaton EM, Gilbert P. 1979. Additivity of action between polysorbate 80 and polymyxin B towards spheroplasts of Pseudomonas aeruginosa NCTC 6750. J Pharm Pharmacol 31:168–170. doi: 10.1111/j.2042-7158.1979.tb13463.x. [DOI] [PubMed] [Google Scholar]
- 107.Landman D, Salamera J, Quale J. 2013. Irreproducible and uninterpretable polymyxin B MICs for Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol 51:4106–4111. doi: 10.1128/JCM.02129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schurek KN, Sampaio JL, Kiffer CR, Sinto S, Mendes CM, Hancock RE. 2009. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:4345–4351. doi: 10.1128/AAC.01267-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hawley JS, Murray CK, Griffith ME, McElmeel ML, Fulcher LC, Hospenthal DR, Jorgensen JH. 2007. Susceptibility of Acinetobacter strains isolated from deployed U.S. military personnel. Antimicrob Agents Chemother 51:376–378. doi: 10.1128/AAC.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matuschek E, Åhman J, Webster C, Kahlmeter G. 2017. Antimicrobial susceptibility testing of colistin—evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin Microbiol Infect 24:865–870. doi: 10.1016/j.cmi.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 111.Tan T, Ng S. 2007. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin Microbiol Infect 13:541–544. doi: 10.1111/j.1469-0691.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 112.Vourli S, Dafopoulou K, Vrioni G, Tsakris A, Pournaras S. 2017. Evaluation of two automated systems for colistin susceptibility testing of carbapenem-resistant Acinetobacter baumannii clinical isolates. J Antimicrob Chemother 72:2528–2530. doi: 10.1093/jac/dkx186. [DOI] [PubMed] [Google Scholar]
- 113.Richter SS, Karichu J, Otiso J, Van Heule H, Keller G, Cober E, Rojas LJ, Hujer AM, Hujer KM, Marshall S, Perez F, Rudin SD, Domitrovic TN, Kaye KS, Salata R, van Duin D, Bonomo RA. 2018. Evaluation of Sensititre broth microdilution plate for determining the susceptibility of carbapenem-resistant Klebsiella pneumoniae to polymyxins. Diagn Microbiol Infect Dis 91:89–92. doi: 10.1016/j.diagmicrobio.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Balouiri M, Sadiki M, Ibnsouda SK. 2016. Methods for in vitro evaluating antimicrobial activity: a review. J Pharm Anal 6:71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 116.Lee SY, Shin JH, Lee K, Joo MY, Park KH, Shin MG, Suh SP, Ryang DW, Kim SH. 2013. Comparison of the Vitek 2, MicroScan, and Etest methods with the agar dilution method in assessing colistin susceptibility of bloodstream isolates of Acinetobacter species from a Korean university hospital. J Clin Microbiol 51:1924–1926. doi: 10.1128/JCM.00427-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li H, Zhou H, Li X, Wang JF, Fu Y, Jiang Y, Yu YS. 2017. The evaluation of four in vitro susceptibility testing methods for colistin on carbapenem-resistant Acinetobacter baumannii. Jundishapur J Microbiol 10:e55956. doi: 10.5812/jjm.55956. [DOI] [Google Scholar]
- 118.Simner PJ, Bergman Y, Trejo M, Roberts AA, Marayan R, Tekle T, Campeau S, Kazmi A, Bell D, Lewis S, Tamma PD, Humphries R, Hindler JA. 3 October 2018. Two-site evaluation of the colistin broth disk elution test to determine colistin in vitro activity against Gram-negative bacilli. J Clin Microbiol doi: 10.1128/JCM.01163-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tam VH, Cao H, Ledesma KR, Hu M. 2011. In vitro potency of various polymyxin B components. Antimicrob Agents Chemother 55:4490–4491. doi: 10.1128/AAC.00119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li J, Turnidge J, Milne R, Nation RL, Coulthard K. 2001. In vitro pharmacodynamic properties of colistin and colistin methanesulfonate against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 45:781–785. doi: 10.1128/AAC.45.3.781-785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Diep JK, Covelli J, Sharma R, Ruszaj DM, Kaye KS, Li J, Straubinger RM, Rao GG. 2018. A comparison of composition and in vitro activity of polymyxin B products. Int J Antimicrob Agents 52:365–371. doi: 10.1016/j.ijantimicag.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 122.Karvanen M, Malmberg C, Lagerback P, Friberg LE, Cars O. 2017. Colistin is extensively lost during standard in vitro experimental conditions. Antimicrob Agents Chemother 61:e00857-17. doi: 10.1128/AAC.00857-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singhal L, Sharma M, Verma S, Kaur R, Britto XB, Kumar SM, Ray P, Gautam V. 2018. Comparative evaluation of broth microdilution with polystyrene and glass-coated plates, agar dilution, E-test, Vitek, and disk diffusion for susceptibility testing of colistin and polymyxin B on carbapenem-resistant clinical isolates of Acinetobacter baumannii. Microb Drug Resist 24:1082–1088. doi: 10.1089/mdr.2017.0251. [DOI] [PubMed] [Google Scholar]
- 124.Figura N, Marcolongo R, Cavallo G, Santucci A, Collodel G, Spreafico A, Moretti E. 2012. Polysorbate 80 and Helicobacter pylori: a microbiological and ultrastructural study. BMC Microbiol 12:217. doi: 10.1186/1471-2180-12-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang H, Shen Y, Weng P, Zhao G, Feng F, Zheng X. 2009. Antimicrobial activity of a food-grade fully dilutable microemulsion against Escherichia coli and Staphylococcus aureus. Int J Food Microbiol 135:211–215. doi: 10.1016/j.ijfoodmicro.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 126.Brown MR, Richards RM. 1964. Effect of polysorbate (Tween) 80 on the resistance of Pseudomonas aeruginosa to chemical inactivation. J Pharm Pharmacol 16:51-5T. [PubMed] [Google Scholar]
- 127.Brown MR, Winsley BE. 1969. Effect of polysorbate 80 on cell leakage and viability of Pseudomonas aeruginosa exposed to rapid changes of pH, temperature and tonicity. J Gen Microbiol 56:99–107. doi: 10.1099/00221287-56-1-99. [DOI] [PubMed] [Google Scholar]
- 128.Sutherland CA, Nicolau DP. 2014. To add or not to add polysorbate 80: impact on colistin MICs for clinical strains of Enterobacteriaceae and Pseudomonas aeruginosa and quality controls. J Clin Microbiol 52:3810. doi: 10.1128/JCM.01454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sei K. 2012. Colistin susceptibility testing and corona treatment January 2012 meeting of the CLSI Subcommittee on Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 130.Aktas G, Derbentli S. 2017. 2016. In vitro activity of daptomycin combined with dalbavancin and linezolid, and dalbavancin with linezolid against MRSA strains. J Antimicrob Chemother 72:441–443. doi: 10.1093/jac/dkw416. [DOI] [PubMed] [Google Scholar]
- 131.Arhin FF, Sarmiento I, Belley A, McKay GA, Draghi DC, Grover P, Sahm DF, Parr TR, Moeck G. 2008. Effect of polysorbate 80 on oritavancin binding to plastic surfaces: implications for susceptibility testing. Antimicrob Agents Chemother 52:1597–1603. doi: 10.1128/AAC.01513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sogaard P, Gahrn-Hansen B. 1986. Population analysis of susceptibility to ciprofloxacin and nalidixic acid in Staphylococcus, Pseudomonas aeruginosa, and Enterobacteriaceae. Acta Pathol Microbiol Immunol Scand B 94:351–356. [DOI] [PubMed] [Google Scholar]
- 133.Pournaras S, Kristo I, Vrioni G, Ikonomidis A, Poulou A, Petropoulou D, Tsakris A. 2010. Characteristics of meropenem heteroresistance in Klebsiella pneumoniae carbapenemase (KPC)-producing clinical isolates of K. pneumoniae. J Clin Microbiol 48:2601–2604. doi: 10.1128/JCM.02134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fernandez-Cuenca F, Gomez-Sanchez M, Rodriguez-Bano J, Martinez-Martinez L, Vila J, Bou G, Bou G, Pascual A. 2012. Epidemiological and clinical features associated with colonisation/infection by Acinetobacter baumannii with phenotypic heterogeneous resistance to carbapenems. Int J Antimicrob Agents 40:235–238. doi: 10.1016/j.ijantimicag.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 135.Hung KH, Wang MC, Huang AH, Yan JJ, Wu JJ. 2012. Heteroresistance to cephalosporins and penicillins in Acinetobacter baumannii. J Clin Microbiol 50:721–726. doi: 10.1128/JCM.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tato M, Morosini M, Garcia L, Alberti S, Coque MT, Canton R. 2010. Carbapenem heteroresistance in VIM-1-producing Klebsiella pneumoniae isolates belonging to the same clone: consequences for routine susceptibility testing. J Clin Microbiol 48:4089–4093. doi: 10.1128/JCM.01130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nakipoglu Y, Derbentli S, Cagatay AA, Katranci H. 2005. Investigation of Staphylococcus strains with heterogeneous resistance to glycopeptides in a Turkish university hospital. BMC Infect Dis 5:31. doi: 10.1186/1471-2334-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morand B, Muhlemann K. 2007. Heteroresistance to penicillin in Streptococcus pneumoniae. Proc Natl Acad Sci U S A 28 104:14098–14103. doi: 10.1073/pnas.0702377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gravey FGJ, Isnard C, Auzou M, Cattoir V, Guérin F. 2017. Screening culture method for reliable detection of colistin heteroresistance in clinical isolates of Enterobacter cloacae complex, poster 1768. Abstr 27th Congr Eur Soc Clin Microbiol Infect Dis (ESCMID), Vienna, Austria. [Google Scholar]
- 140.Charretier Y, Diene SM, Baud D, Chatellier S, Santiago-Allexant E, van Belkum A, Guigon G, Schrenzel J. 2018. Colistin heteroresistance and the involvement of the PmrAB regulatory system in Acinetobacter baumannii. Antimicrob Agents Chemother 62:e00788-18. doi: 10.1128/AAC.00788-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fernandez-Mazarrasa C, Mazarrasa O, Calvo J, del Arco A, Martinez-Martinez L. 2009. High concentrations of manganese in Mueller-Hinton agar increase MICs of tigecycline determined by Etest. J Clin Microbiol 47:827–829. doi: 10.1128/JCM.02464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Girardello R, Bispo PJ, Yamanaka TM, Gales AC. 2012. Cation concentration variability of four distinct Mueller-Hinton agar brands influences polymyxin B susceptibility results. J Clin Microbiol 50:2414–2418. doi: 10.1128/JCM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Davis SD, Iannetta A, Wedgwood RJ. 1971. Activity of colistin against Pseudomonas aeruginosa: inhibition by calcium. J Infect Dis 124:610–612. [DOI] [PubMed] [Google Scholar]
- 144.Felegie TP, Yu VL, Rumans LW, Yee RB. 1979. Susceptibility of Pseudomonas maltophilia to antimicrobial agents, singly and in combination. Antimicrob Agents Chemother 16:833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Matzneller P, Strommer S, Osterreicher Z, Mitteregger D, Zeitlinger M. 2015. Target site antimicrobial activity of colistin might be misestimated if tested in conventional growth media. Eur J Clin Microbiol Infect Dis 34:1989–1994. doi: 10.1007/s10096-015-2441-7. [DOI] [PubMed] [Google Scholar]
- 146.Andrews JM. 2001. The development of the BSAC standardized method of disc diffusion testing. J Antimicrob Chemother 48:29–42. doi: 10.1093/jac/48.suppl_1.29. [DOI] [PubMed] [Google Scholar]
- 147.Andrews J, Walker R, King A. 2002. Evaluation of media available for testing the susceptibility of Pseudomonas aeruginosa by BSAC methodology. J Antimicrob Chemother 50:479–486. [DOI] [PubMed] [Google Scholar]
- 148.Andrews J. 2007. BSAC standardized disc susceptibility testing method (version 6). J Antimicrob Chemother 60:20–41. doi: 10.1093/jac/dkm110. [DOI] [PubMed] [Google Scholar]
- 149.Gwozdzinski K, Azarderakhsh S, Imirzalioglu C, Falgenhauer L, Chakraborty T. 2018. An improved medium for colistin susceptibility testing. J Clin Microbiol 56:e01950-17. doi: 10.1128/JCM.01950-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lee J-Y, Park YK, Chung ES, Na IY, Ko KS. 2016. Evolved resistance to colistin and its loss due to genetic reversion in Pseudomonas aeruginosa. Sci Rep 6:25543. doi: 10.1038/srep25543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gomez-Simmonds A, Uhlemann A-C. 2017. Clinical implications of genomic adaptation and evolution of carbapenem-resistant Klebsiella pneumoniae. J Infect Dis 215:S18–S27. doi: 10.1093/infdis/jiw378. [DOI] [PMC free article] [PubMed] [Google Scholar]