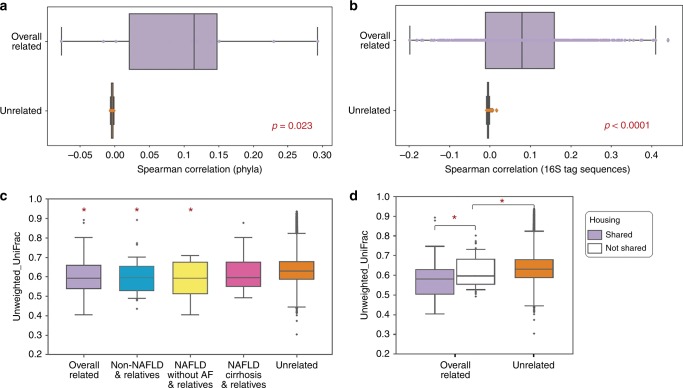
Fig. 1.
Familial association and shared microbiome among relatives is driven by shared housing. Distribution of spearman correlation coefficients between relatives in purple (n = 86) and unrelated pairs in orange (n = 18232) in the familial cohort, plotted for each phyla (a) and 16S tag sequences (b). The box plots show the quartiles and whiskers show the rest of the distribution (1.5 inter-quartile range) and the center line corresponds to the median. This analysis was done after filtering rare 16S sequences to avoid spurious correlations due to sparsity (total abundance <10E-6 across all samples in each disease group). The correlation among related individuals was significantly higher at both phylum (p = 0.023) and 16S tag sequences (p = 2.4E-41) levels. Similar plot showing the distribution of unweighted UniFrac distances between related in purple and unrelated pairs stratified by disease status (c). The beta-diversity was significantly lower among related individuals (p = 3.22E-05), non-NAFLD controls and relative in blue (n = 38 pairs) (p = 0.0011) and probands with NAFLD without AF and relatives in yellow (n = 15) (p = 0.0156) when compared to the same among unrelated pairs in orange, while the difference between NALFD-cirrhosis patients and relatives in pink (n = 33) and unrelated pairs in orange was not statistically significant (p > 0.1). When stratified by shared housing (d), beta-diversity was significantly lower among related individuals sharing a house in purple (n = 35 pairs) (p = 0.0455). Additionally, related individuals not sharing a house in white (n = 51 pairs) had significantly lower beta-diversity compared to unrelated pairs in orange (p = 0.028). All p value were determined by two-sided Kruskal-Wallis test. *p value < 0.05. Source data are provided as a Source Data file