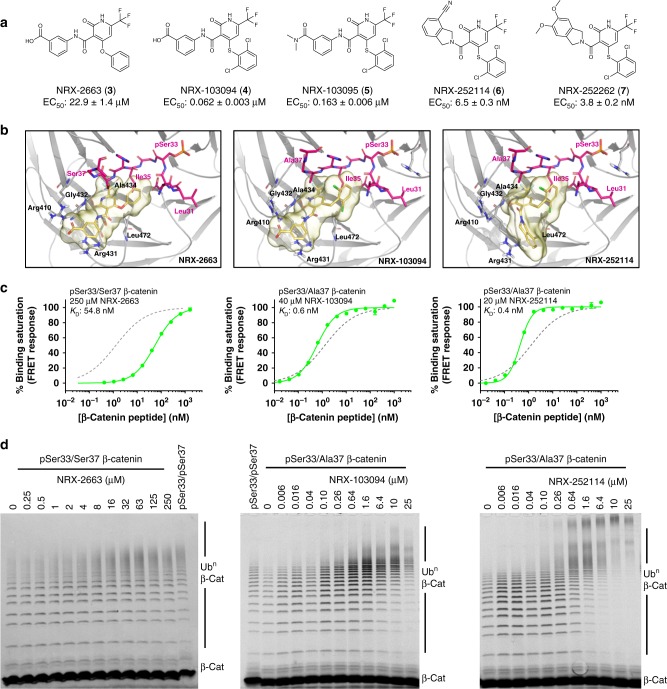
Fig. 3.
Optimized enhancers potentiate pSer33/S37A β-catenin binding and ubiquitylation. a Chemical structure of optimized enhancers and their potency in TR-FRET assay against pSer33/S37A β-catenin peptide for β-TrCP binding. b Structures of enhancers (yellow) in complex with β-catenin peptide (magenta) and β-TrCP (gray). The binding poses and β-catenin degron conformations shown for NRX-2663 (PDB: 6M92) and NRX-103094 (6M91) are from crystal structures (Supplementary Table 1), whereas the NRX-252114 pose was determined by computational docking. c Binding affinities of the β-catenin peptides (as indicated) for β-TrCP were measured in the TR-FRET assay in presence of enhancers at concentration indicated. For NRX-252114, the binding assay is bottoming out due to 300 pM β-TrCP concentration, with the curve fit yielding a higher than expected hill slope of 1.8. Therefore, the KD reported for NRX-252114 represents an upper estimate. For reference, the binding curve for pSer33/pSer37 β-catenin peptide in the same TR-FRET assay is represented in dotted gray line with a KD of 2 nM. d 4 μM of fluorescently-labeled β-catenin peptide (residues 17–60; as indicated) was ubiquitylated in the presence of 125 nM Ube1, ATP, 1.75 μM Cdc34 and 100 nM SCFβ-TrCP with increasing concentration of enhancers. β-Catenin ubiquitylation were resolved by SDS-PAGE and imaged for fluorescence