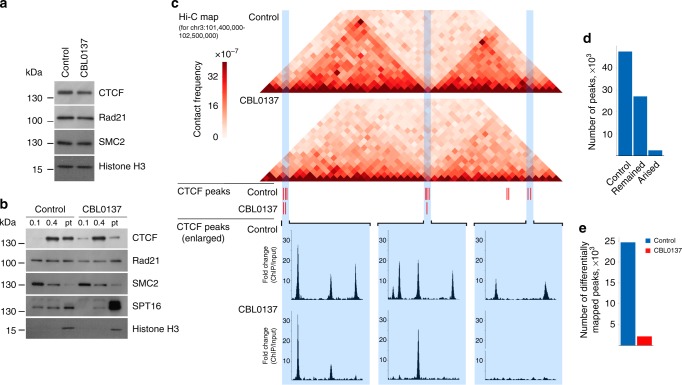
Fig. 6.
CBL0137 induces partial dissociation of CTCF from its binding sites. a Western blot analysis of CTCF, Rad21 (cohesin subunit), and SMC2 (condensin subunit) in nuclear extracts prepared from control and CBL0137-treated (3 µM, 6 h) HT1080 cells. Histone H3 was used as a loading control. b Western blot analysis of CTCF, Rad21, SMC2, SPT16 (FACT subunit), and histone H3 in different chromatin fractions from control and CBL0137-treated (3 µM, 6 h) HT1080 cells prepared by sequential extraction with 0.1 and 0.4 M NaCl. Pellet (Pt): insoluble chromatin fraction. c Representative 1.1 Mb-long genomic segment from chromosome 3 containing two distinct chromatin loop domains bordered by CTCF binding sites in control cells. Corresponding Hi-C maps, CTCF peaks determined using the PePr computational approach[43], and an enlarged view of the particular groups of CTCF peaks are shown for control and CBL0137-treated HT1080 cells. d Bar plots showing the total number of CTCF peaks mapped by PePr in intact HT1080 cells, preserved after CBL0137 treatment, and the newly formed CTCF peaks in CBL0137-treated cells. e Bar plots showing the number of CTCF differential binding sites identified in control and CBL0137-treated HT1080 cells. Analysis of differential binding sites was performed using PePr43, which showed the number of peaks mapped only in one of the two samples (either control or CBL0137-treated). Source data of Fig. 6a, b are provided in a Source Data file