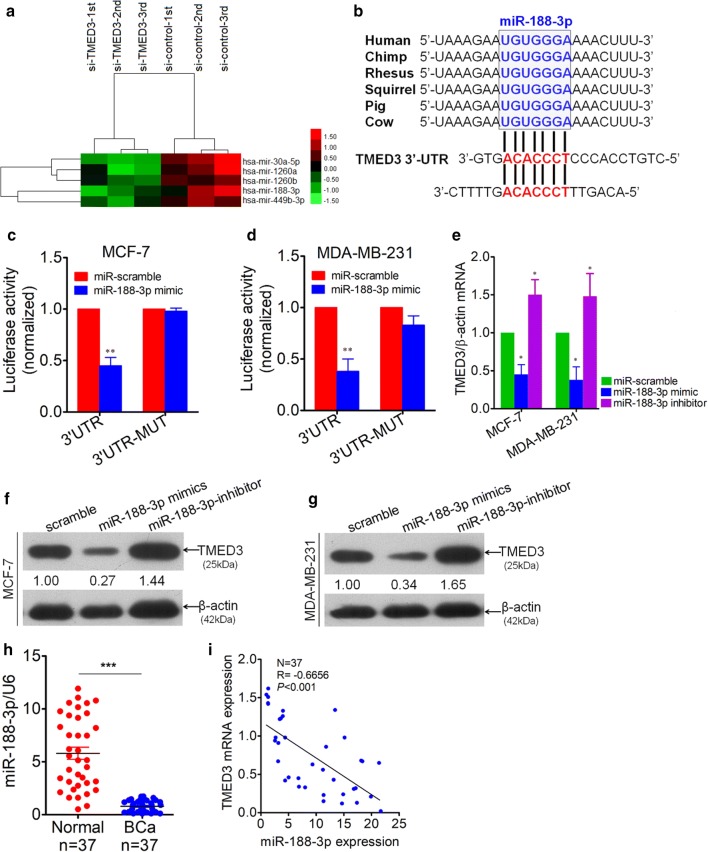
Fig. 4.
TMED3 was identified as a down-stream target of miR-188-3p. a Most significantly up-regulated miRNAs, totaling 5, were screened out using miRNA microarray analysis between group transfected with si-TMED3 and control group, with each group in triplicate. b Conservation of miR-188-3p among mammals by Targetscan (Targetscan Human 7.2 version released) and two potential binding sites (in red) were analyzed in 3′-UTR of TMED3. c Double Luciferase reporter assay was performed between miR-188-3p and 3′-UTR of TMED3 in MCF-7 cells. **p < 0.01 compared with control using independent sample T-test. d Likewise, Luciferase reporter assay was performed in parallel in MDA-MB-231 cells. **p < 0.01 compared with control using independent sample T-test. e Variation of TMED3 mRNA was detected by qRT-PCR after transfection with miR-188-3p mimics and inhibitor sequence in MCF-7 and MDA-MB-231 cells, *p < 0.05 compared with control group. Shown were the representative figures selected among independent three times of repeats. f, g Variation of TMED3 on protein level was evaluated by western-blot, presented was the representative figure picked out from three times of independent repeat. Densitometric analysis of TMED3 blots were performed using Image J software (NIH, Bethesda, USA). h Detection of miR-188-3p by qRT-PCR in breast cancer (BCa) tissues and its matched normal controls, totaling 37. U6, as internal loading control; ***p < 0.001 compared with normal control using independent sample T-test; i correlation between miR-188-3p and TMED3 expression level was analyzed using Pearson correlation test in 37 cases of BCa; r = − 0.6656, p < 0.001 using Pearson Correlation test