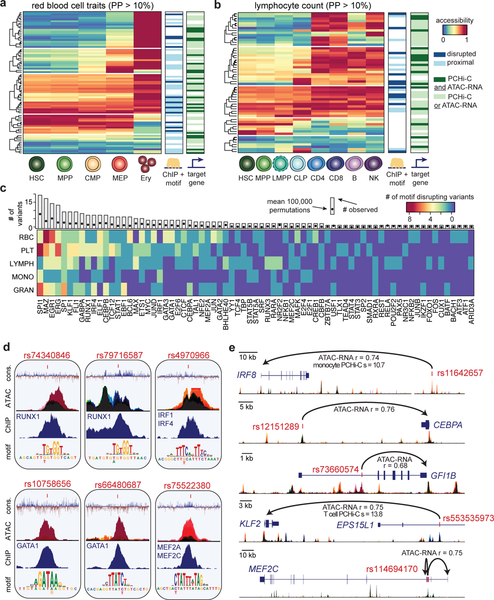
Figure 2 |. Mechanisms of core gene regulation in blood production.
(a,b) Heatmaps depicting red blood cell trait-associated variants (PP > 0.10) across the erythroid lineage (a) and lymphocyte count-associated variants (PP > 0.10) across the lymphoid lineage (b), clustered by chromatin accessibility. Each row marks a fine-mapped variant, each column denotes a cell type within the relevant lineage, and color denotes relative chromatin accessibility along the lineage at each variant (blue, least accessible chromatin; red, most accessible chromatin). Putative target genes (predicted by ATAC-RNA correlation and/or PCHi-C) and disrupted TFs (predicted by ChIP-seq occupancy and motif disruption) are indicated to the right. (c) Transcription factor motifs disrupted in lineage-specific hematopoietic traits. Each row represents a set of traits where variants disrupt specified TF motifs and are occupied by that TF in hematopoietic cells. The unique margin sums across each lineage are shown in the bar plot for each TF. The expected number of variants with ChIP + motif disruption across all PPs is estimated using 100,000 permutations and is shown as a single point. (d) Examples of molecular mechanisms from the analysis in c reveals putative causal variants that disrupt cis-binding of hematopoietic TFs known to be involved in regulating hematopoiesis for various blood cell traits: rs10758656 and rs66480687 are associated with red blood cell traits; rs75522380 and rs74340846 are associated with platelet traits; rs4970966 is associated with monocyte count; and rs79716587 is associated with lymphocyte count. Black color represents accessibility throughout hematopoiesis, whereas other stacked colors represent accessibility for the cell types shown in Figure 3d. (e) Examples of putative target genes from the analysis in a and b: rs11642657 and rs12151289 are associated with monocyte count; rs73660574 is associated with red blood cell traits; rs553535973 is associated with lymphocyte count; and rs114694170 is associated with platelet traits. Colors for accessible chromatin are the same as in d.