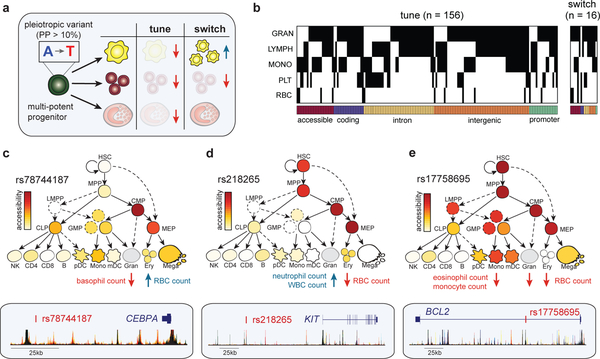
Figure 4 |. Dissecting mechanisms of pleiotropic variants across multiple blood cell lineages.
(a) Schematic that illustrates fine-mapped variants acting in multi-potential or heterogeneous progenitors on distinct hematopoietic lineages, either by tuning lineages in the same direction or switching the regulation in opposite directions. (b) A heatmap depicting 172 fine-mapped variants (PP > 0.10) with pleiotropic effects on cell counts in two or more hematopoietic lineages (eosinophil, neutrophil, basophil, lymphocyte, monocyte, platelet, RBC). Effects on eosinophil, neutrophil, and basophil counts are visualized together as a singular granulocyte lineage. Genomic annotations are indicated below each variant. (c) Pleiotropic variant rs78744187, located downstream of CEBPA, has high chromatin accessibility in CMP and MEP progenitors (top) and demonstrates a switch mechanism by downregulating basophil count while upregulating RBC count (bottom). (d) rs218265, located upstream of stem cell factor KIT, has high chromatin accessibility in several early progenitors (HSC, MPP, CMP, MEP) and demonstrates a switch mechanism by upregulating neutrophil and WBC count while downregulating RBC count. (e) rs17758695, located within an intron of anti-apoptotic factor BCL2, has high chromatin accessibility in several early progenitors (HSC, MPP, CMP, MEP) and exhibits a tuning mechanism, simultaneously downregulating eosinophil, monocyte, and RBC counts.