Abstract
Proteases from flaviviruses have gained substantial interest as potential drug targets to combat infectious diseases caused by dengue, West Nile, Zika and related viruses. Despite nearly two decades of drug discovery campaigns, promising lead compounds for clinical trials have not yet been identified. The main challenges for successful lead compound development are associated with limited drug-likeness of inhibitors and structural ambiguity of the protease target. This brief review focuses on the available information on the structure of flavivirus proteases and their interactions with inhibitors and attempts to point the way forward for successful identification of future lead compounds.
Keywords: Flavivirus, Serine protease, NS2B-NS3, Protease inhibitor
Introduction
The genus flavivirus includes over 50 viruses and belongs to the family of Flaviviridae (Barrows et al. 2018). Each year, millions of people worldwide are infected by members of the flavivirus genus (Boldescu et al. 2017). Some like yellow fever, Japanese encephalitis or tick-borne encephalitis can be prevented through the use of established vaccines. Others like dengue, West Nile or Zika represent major health burdens without available specific antiviral treatments or effective and safe vaccines. Flaviviruses are usually vector-transmitted (e.g. by the mosquitos Aedes aegypti and Aedes albopictus), restricting their distribution to the continuously expanding vector habitats. Additional transmission routes for Zika virus involving sexual contact have recently been proposed (Baud et al. 2017; Poland et al. 2018).
Estimates of the annual dengue virus infections are between 284 and 528 million, of which between 67 and 136 million cases manifest clinically (Bhatt et al. 2013). The efficacy of the recently approved vaccine CYD-TDV (Dengvaxia) differs amongst the four known dengue serotypes and age groups of those receiving the vaccine. In addition, the vaccine performs differently in individuals with evidence of prior dengue infection (seropositive) and those without (seronegative), with an increased risk for hospitalisation in the latter group (WHO 2018). Consequently, the WHO recommends the current vaccine only for seropositive patients.
West Nile virus affects animals (e.g. birds and horses) and humans (Suthar et al. 2013). Approximately 80% of human West Nile virus infections are asymptomatic, 20% cause self-limiting symptoms (West Nile fever) and less than 1% are characterised by neuroinvasive disease, with 10% of this subgroup resulting in fatality (Burki 2018). Although candidates are in clinical trials, no West Nile virus vaccine for humans has yet been approved. During 2018, Europe registered an alarming increase of West Nile cases with more than 2000 reports of autochthonous infections and 180 deaths, exceeding the total number of cases reported during the previous 7 years (ECDC 2018).
The Zika virus has emerged very recently as a health-threatening pathogen after epidemic outbreaks in Latin America (Baud et al. 2017). Most infections are asymptomatic; however, neurological complications in patients and severe fetal disorders (microcephaly) prompted the WHO to declare Zika virus a Public Health Emergency of International Concern in 2016 (Baud et al. 2017). Since that time, several vaccine candidates have been developed, of which four are currently in clinical trials (Poland et al. 2018). Potential cross-reactions between Zika and dengue virus antibodies (antibody-dependent enhancements) may lead to increased viremia and severity of the disease and thus challenge vaccination campaigns where dengue and Zika co-circulate (Poland et al. 2018).
These examples describing the challenges of vaccine development illustrate that anti-flaviviral drugs must be pursued in addition to vaccination campaigns to present therapeutic options for the treatment of symptomatic patients and individuals where vaccination is not recommended (e.g. dengue seronegative individuals). In what follows, the current state of play in targeting the protease of flaviviruses is described.
Function of the NS2B-NS3 protease
All members of the flavivirus genus contain a single-stranded RNA genome that is translated by the host cell into a single polyprotein (Barrows et al. 2018). Embedded into the membrane of the endoplasmatic reticulum, this precursor protein is post-translationally processed by host cell proteases and the viral protease NS2B-NS3 into three structural and various non-structural (NS) proteins (Barrows et al. 2018; Boldescu et al. 2017). Consequently, the NS2B-NS3 protease is essential for viral replication and thus presents itself as a promising drug target (Nitsche 2018; Nitsche et al. 2014). The catalytically active protease complex consists of the actual protease unit located at the N-terminal part of the non-structural protein 3 (NS3) and requires approximately 40 residues of a hydrophilic domain of the membrane-associated non-structural protein 2B (NS2B) as a cofactor. NS2B-NS3 is a serine protease with the highly conserved catalytic triad (serine 135, histidine 51 and aspartate 75) (Nitsche et al. 2014). According to the nomenclature for protease subsites suggested by Schechter and Berger (1967), the NS2B-NS3 proteases show a common preference to cleave peptidic backbones after two basic residues (arginine or lysine) in P1 and P2. In contrast to other viral protease drug targets that tolerate more hydrophobic substrate residues in these key positions (e.g. HIV, HCV), this dibasic recognition motif has manifested as a major challenge to the drug discovery process against flaviviruses.
Constructs for drug discovery
Protease constructs with full-length NS2B that rely on lipid support (e.g. micelles) have been reported (Choksupmanee et al. 2012; Huang et al. 2013; Ng et al. 2019), but the most commonly used constructs for biochemical and structural studies comprise only the hydrophilic core segment of NS2B, excluding the hydrophobic membrane-associated domains. These truncated constructs have been designed with and without covalent linkage between NS2B and NS3 (Nitsche et al. 2014). The most frequently used truncated construct includes a covalent Gly4-Ser-Gly4 linker between NS2B and NS3, as initially introduced for dengue serotype 2 (Leung et al. 2001), followed by West Nile (Nall et al. 2004), the other dengue serotypes (Li et al. 2005) and most recently Zika virus protease (Lei et al. 2016). Unlinked constructs rely either on an NS2B/NS3 autocleavage site (de la Cruz et al. 2014; Phoo et al. 2016) or co-expression systems (Kim et al. 2013; Zhang et al. 2016). There is an ongoing debate in the literature whether the linked or unlinked protease constructs are more suitable for drug discovery campaigns (Kang et al. 2017; Nitsche 2018; Nitsche et al. 2014). The linked constructs tend to be more stable, whereas the unlinked constructs are thought to resemble a state that is closer to the native one (de la Cruz et al. 2014; Hill et al. 2018; Li et al. 2017a). Recent studies with Zika virus protease suggest that the covalent linker may prevent the formation of the catalytically active complex to a certain degree (Li et al. 2017a), whereas no significant differences for inhibitor binding could be observed between linked and unlinked Zika virus protease constructs (Nitsche et al. 2019).
Structure of the NS2B-NS3 protease
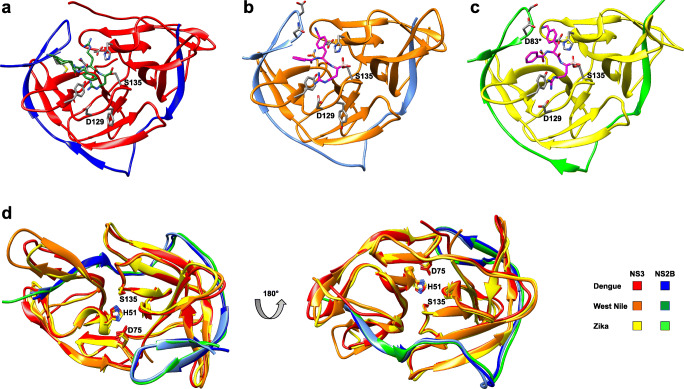
Figure 1 shows the NS2B-NS3 proteases from dengue, West Nile and Zika viruses in their catalytically active form (Lei et al. 2016; Nitsche et al. 2017; Noble et al. 2012). These structures were produced by co-crystallising substrate-derived ligands in the active site. In case of dengue, an active structure could so far only be solved for serotype 3 (Noble et al. 2012). These protease structures share a high degree of similarity with several conserved structural features, as illustrated by superimposition (Fig. 1d). This has raised hopes that once a promising lead compound is discovered, it could be considered as a potential pan-flaviviral protease inhibitor to treat dengue, West Nile, Zika and related flaviviral infectious diseases. In its active form, the protease adopts a chymotrypsin-like fold where the N-terminal β-strand of NS2B integrates into the N-terminal domain of NS3 and the C-terminal β-hairpin of NS2B wraps around the active site of NS3. The latter interaction forms the S2 pocket and is thus essential for substrate recognition and catalytic activity. Only the protease from Zika virus can engage in a salt bridge between the P2 side chain and the NS2B aspartate residue D83* (Fig. 1c), which is not conserved in West Nile and all four dengue serotypes. It has been suggested that this additional ionic interaction between substrate (P2) and NS2B (S2) is partially responsible for the observed hyperactivity of Zika virus protease (Lei et al. 2016). A second substantial interaction involves the highly conserved NS3 aspartate residue D129 and the basic P1 substrate side chain, which form a salt bridge in all structures (Fig. 1a–c).
Fig. 1.
Ligand-bound X-ray co-crystal structures of active flavivirus NS2B-NS3 proteases. (a) Dengue protease serotype 3 with the covalently bound aldehyde ligand 1 (3U1I) (Noble et al. 2012). (b) West Nile protease with the covalently bound boronate ligand 2 (5IDK) (Nitsche et al. 2017). (c) Zika protease with the covalently bound boronate ligand 2 (5LC0) (Lei et al. 2016). (d) Superimposition of active conformations of NS2B-NS3 proteases from dengue (3U1I), West Nile (5IDK) and Zika (5LC0) viruses. Co-crystallised ligands have been removed from the structures. Residues of the catalytic triad are indicated. Residues marked with an asterisk indicated NS2B. This figure has been generated with Chimera (Pettersen et al. 2004)
Open or closed?
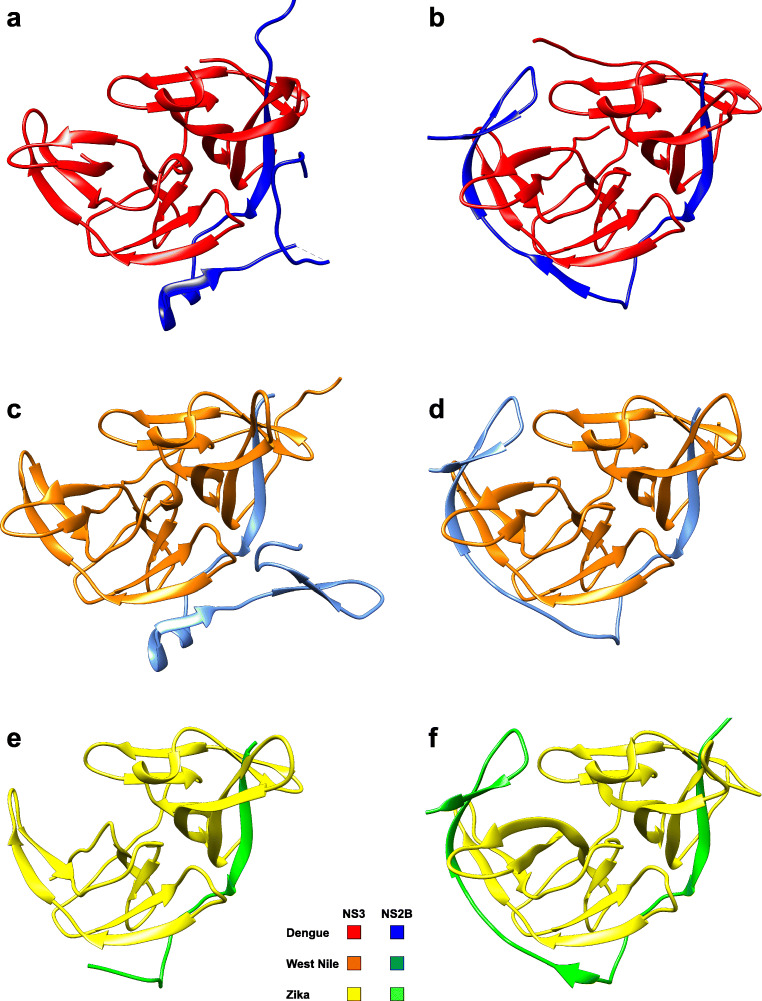
Two major groups of X-ray crystal structures have been solved for each viral protease (dengue, West Nile, Zika) that differ substantially in the C-terminal NS2B domain (Fig. 2). One group (crystallised in the absence of inhibitor) observes the C-terminal part of NS2B, either dissociated from NS3, or not defined by electron density (Fig. 2a, c, e). The second group (usually crystallised in the presence of an inhibitor) defines the C-terminal NS2B domain as a β-hairpin wrapped around the active site (Fig. 2b, d, f). The former inactive state is often referred to as open, whereas the latter active one is known as closed. These crystallographic observations indicated substantial conformational changes of NS2B upon activation by substrate or inhibitor. Particularly, the early unliganded open crystal structures from dengue (Erbel et al. 2006) and West Nile (Aleshin et al. 2007) proteases imply this conclusion. However, recent observations of a crystal structure of Zika virus protease in the closed conformation were made for the unlinked protease construct in the absence of a ligand (Zhang et al. 2016). In contrast, unliganded structures from linked constructs of dengue (Erbel et al. 2006), West Nile (Aleshin et al. 2007) and Zika (Chen et al. 2016) proteases all revealed inactive open states, suggesting that the covalent Gly4SerGly4 linker between NS2B and NS3 prevents the formation of the closed conformation in the absence of substrate or inhibitor.
Fig. 2.
X-ray crystal structures of inactive (open) and active (closed) conformations of flavivirus NS2B-NS3 proteases. Co-crystallised ligands have been removed from the active (closed) structures. (a) Inactive (open) conformation of the dengue protease serotype 2 (2FOM) (Erbel et al. 2006). (b) Active (closed) conformation of the dengue protease serotype 3 (3U1I) (Noble et al. 2012). (c) Inactive (open) conformation of the West Nile protease mutant H51A (2GGV) (Aleshin et al. 2007). (d) Active (closed) conformation of the West Nile protease (5IDK) (Nitsche et al. 2017). (e) Inactive (open) conformation of the Zika protease (5GXJ) (Chen et al. 2016). (f) Active (closed) conformation of the Zika protease (5LC0) (Lei et al. 2016). This figure has been generated with Chimera (Pettersen et al. 2004)
Various biomolecular NMR studies were conducted to analyse the conformational ambiguity of flavivirus proteases in solution, particularly with respect to the localisation of the C-terminal part of NS2B. After initial challenges with heterogeneity in sample preparations, most reports concluded that both, the dengue and West Nile virus proteases, predominately adopt the closed conformation in solution, regardless of the construct (linked or unlinked) or the presence of an inhibitor (de la Cruz et al. 2014, 2011; Kim et al. 2013; Su et al. 2009). Observed conformational flexibilities may relate to exchange phenomena between open and closed states, the extent of which can differ between constructs (Hill et al. 2018). In the case of Zika virus protease, NMR experiments indicated sample heterogeneity promoted by the Gly4SerGly4 linker and ligand-binding events (Li et al. 2017a; Mahawaththa et al. 2017). Distances measured by double electron-electron resonance in the linked construct of the Zika virus protease matched the closed conformation in presence and absence of a ligand (Mahawaththa et al. 2018). In case of the unlinked Zika virus protease constructs, the closed conformation was found to be dominant in solution with a tendency of additional stabilisation in the presence of a ligand (Li et al. 2018, 2017b; Phoo et al. 2016; Zhang et al. 2016).
Most importantly, all studies showed that regardless of the protease construct, the enzymes are folded correctly in solution. Ambiguities are mainly attributed to partial dissociations between NS2B and NS3 as well as inhibitor-binding events. A certain degree of conformational flexibility of NS2B-NS3, particularly in the C-terminal region of NS2B is not surprising, as the substrate must enter the active site prior cleavage reaction. Substrate binding stabilises the active conformation but is not an essential requirement to assemble the active closed NS2B-NS3 complex. Therefore, the closed state should be the preferred template for rational drug discovery attempts. There is a general tendency that the unlinked constructs resemble the closed and active state more reliably than the linked variants.
Inhibitors
Several inhibitors of flaviviral proteases have been described and comprehensively reviewed (Behnam et al. 2016; Boldescu et al. 2017; Lim and Shi 2013; Lim et al. 2013; Nitsche 2018; Nitsche et al. 2014; Poulsen et al. 2014; Timiri et al. 2016). They can generally be categorised as either substrate-derived peptides/peptidomimetics or small molecules without substrate character (Nitsche 2018). The former group is usually characterised by high affinity and limited drug-likeness, whereas the latter group implies the opposite properties: improved drug-likeness, but lower affinity. Only few inhibitors have been reported that display dissociation constants with flaviviral proteases in the desirable lower nanomolar range, i.e. clearly below 100 nM (Behnam et al. 2015; Nitsche et al. 2017; Schüller et al. 2011; Shiryaev et al. 2006; Stoermer et al. 2008; Yin et al. 2006) and all of them are peptidic-based substrate mimetics with the majority bearing warheads that allow covalent modification of the catalytically active serine residue 135.
Most campaigns focused on the discovery of competitive inhibitors that bind to the active site. As a consequence, the undesirable dibasic substrate recognition motif is reflected in several compounds. Out of those inhibitors that have been proven to bind to the active site by X-ray crystallography (Fig. 3), only two are non-peptidic and do not comprise at least two basic side chains. Only a few attempts were made to discover allosteric inhibitors that do not interact with the active site (Brecher et al. 2017; Nitsche et al. 2019; Roy et al. 2017; Shiryaev et al. 2017; Wu et al. 2015; Yildiz et al. 2013). To date, a distinct binding mode has not been reported for any. Once structural data will become available, more rational attempts towards allosteric inhibitors can be pursued.
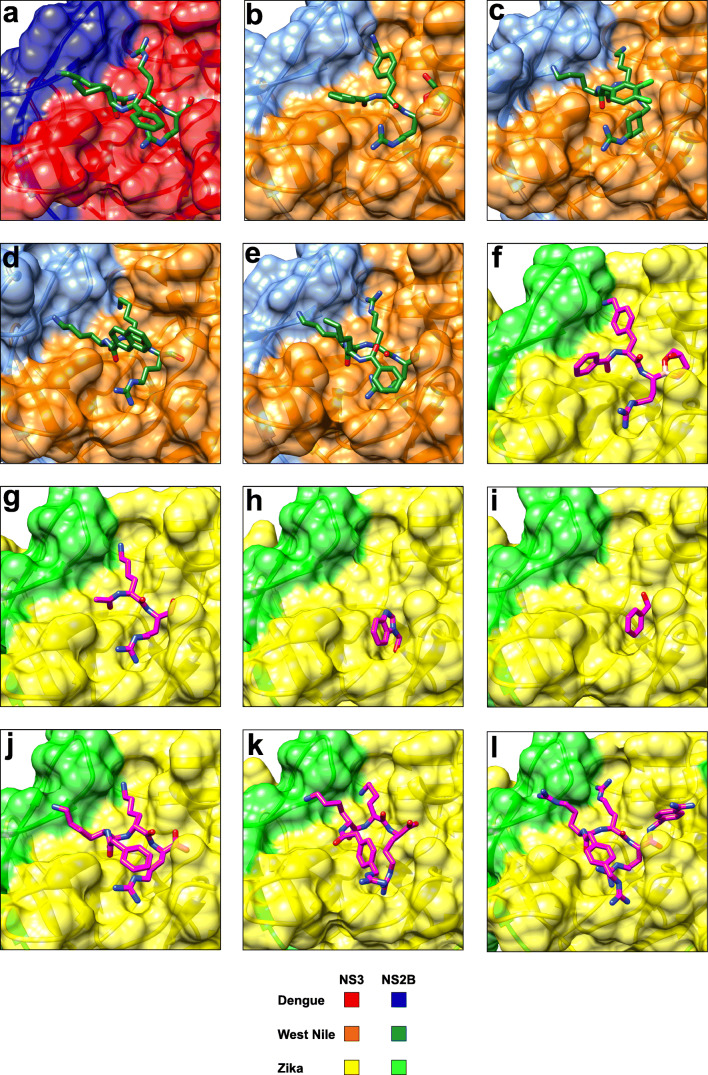
Fig. 3.
X-ray co-crystal structures of flavivirus NS2B-NS3 proteases in complex with active-site inhibitors. (a) Dengue protease serotype 3 in complex with compound 1 (3U1I) (Noble et al. 2012). The aldehyde in 1 forms a covalent hemiacetal with S135 (not shown). (b) West Nile protease in complex with compound 2 (5IDK) (Nitsche et al. 2017). The boronic acid in 2 forms a cyclic ester adduct with glycerol (as shown) and a covalent boronate with S135 (not shown). (c) West Nile protease in complex with compound 3 (2YOL) (Hammamy et al. 2013). (d) West Nile protease in complex with compound 4 (3E90) (Robin et al. 2009). The aldehyde in 4 forms a covalent hemiacetal with S135 (not shown). (e) West Nile protease in complex with compound 1 (2FP7) (Erbel et al. 2006). The aldehyde in 1 forms a covalent hemiacetal with S135 (not shown). (f) Zika protease in complex with compound 2 (5LC0) (Lei et al. 2016). The boronic acid in 2 forms a cyclic ester adduct with glycerol (as shown) and a covalent boronate with S135 (not shown). (g) Zika protease C143S mutant in complex with compound 5 (5YOF) (Li et al. 2018). The aldehyde in 5 forms a covalent hemiacetal with S135 (not shown). A similar structure of lower resolution has also been reported for compound 5 in complex with the Zika protease wildtype (5H6V) (Li et al. 2017b). (h) Zika protease in complex with fragment 6 (5H4I) (Zhang et al. 2016). (i) Zika protease in complex with a benzoyl fragment (5YOD) (Li et al. 2018). Transesterification between Zika protease and compound 7 results in the formation of a S135 benzoate (covalent bond not shown). (j) Zika protease C143S mutant in complex with proteolytically cleaved compound 8 (5ZMQ) (Phoo et al. 2018). (k) Zika protease C143S mutant in complex with proteolytically hydrolysed compound 9 (5ZMS) (Phoo et al. 2018). (l) Zika protease C143S mutant in complex with compound 10 (5ZOB) (Phoo et al. 2018). This figure has been generated with Chimera (Pettersen et al. 2004)
Co-crystal structures of NS2B-NS3 and inhibitors
Over the past 3 years, massive efforts in X-ray crystallography have generated substantial structural information about the activation of flavivirus proteases and their interactions with inhibitors.
Many crystal structures of dengue virus NS2B-NS3 protease have been solved for serotypes 1 (3L6P, 3LKW) (Chandramouli et al. 2010), 2 (2FOM, 4M9K, 4M9M, 4M9I, 4M9F, 4M9T) (Erbel et al. 2006; Yildiz et al. 2013), 3 (3U1I, 3U1J) (Noble et al. 2012) and for the full-length protease-helicase complex of serotype 4 (2VBC, 2WHX, 2WZQ) (Luo et al. 2010, 2008). Recently, additional NS2B-NS3 structures with full-length NS3 (protease and helicase) of serotype 4 have been deposited in the protein data base (5YVU, 5YVJ, 5YVW, 5YVY, 5YW1). Two structures were solved in complex with the 6.5 kDa broad-spectrum protease inhibitor aprotinin (also referred to as BPTI) for serotypes 3 (3U1J) (Noble et al. 2012) and 4 (5YW1). Despite the large total number of available crystal structures, only one describes a small molecule inhibitor (1) in complex with the dengue serotype 3 protease (Fig. 3a) (Noble et al. 2012). Tetrapeptide 1 resembles the non-prime site of the substrate recognition sequence from P1 to P4 (Nle-Lys-Arg-Arg), which was previously identified from a large screening campaign (Li et al. 2005). An additional aldehyde function at the peptide C-terminus allows covalent linkage to serine 135.
In stark contrast to dengue, the majority of crystal structures of the West Nile virus protease have been solved in complex with inhibitors. One unliganded (2GGV) and one structure in complex with aprotinin (2IJO) have been reported (Aleshin et al. 2007), all others feature small molecules (Fig. 3b–e). Compound 2 represents a short peptide mimetic that binds covalently to the catalytic serine 135 via a boronic acid warhead (Fig. 3b), while its two basic side chains occupy the S1 and S2 pockets (Nitsche et al. 2017). Compound 3 is the only co-crystallised small molecule that does not covalently modify the West Nile virus protease (Fig. 3c) (Hammamy et al. 2013). Similar to the complex of dengue protease with compound 1, the three basic side chains of inhibitor 3 occupy S1-S3, which is a common feature in several structures of flavivirus proteases in complex with peptide inhibitors (e.g. Fig 3a, c, d, e, j, k, l). Compounds 4 and 1 are tri- and tetrapeptide aldehydes, respectively, that bind covalently to serine 135 (Fig. 3d, e) (Erbel et al. 2006; Robin et al. 2009). The binding mode of compound 1 to West Nile protease (Fig. 3e) is similar to the interaction of 1 with dengue protease serotype 3 (Fig. 3a).
Recently, several crystal structures of Zika virus protease were reported, of which some lack ligands, like 5GXJ (Chen et al. 2016), 5GPI (Zhang et al. 2016), 5T1V, 5TFN and 5TFO. Others revealed active-site complexes with terminal peptide chains from NS2B (5GJ4) (Phoo et al. 2016) or NS3 5GPI (Zhang et al. 2016). Additional structures have been solved in complex with small molecule inhibitors bound to the active site (Fig. 3f–l). The first crystal structure became available in complex with compound 2 covalently linked to S135 (Lei et al. 2016). The binding mode of 2 is similar to that one observed in complex with West Nile virus protease (Fig. 1b, Fig. 3b), except the additional salt bridge between P2 and D83* of NS2B (Fig. 1c, Fig. 3f). Compound 5 is a short dipeptide aldehyde that binds covalently to S135 and occupies S1 and S2 with its two basic side chains (Fig. 3g) (Li et al. 2018, 2017b) similarly to boronate 2. The low-affinity fragment 6 represents the first very small molecule co-crystallised with a flavivirus protease (Fig. 3h) (Zhang et al. 2016). It is involved in stacking interactions with Y161 and hydrogen bonding with Y150 in the S1 pocket without any contacts to NS2B. Activated pyrazole ester 7 and analogues are known to inhibit the proteases from dengue, West Nile and Zika viruses by transesterification (Johnston et al. 2007; Koh-Stenta et al. 2015; Li et al. 2018; Sidique et al. 2009). In the final inhibition state, the benzoate is covalently linked to S135, engaging in π-π stacking with Y161 and hydrogen-bonding with H51 in the S1 pocket (Fig. 3i) (Li et al. 2018). Finally, the peptidic inhibitors 8–10 have been co-crystallised with Zika virus protease (Fig. 3j-l) (Phoo et al. 2018). Only compound 10 was found be resistant to proteolysis. With two glycines in P1’ and P2’, compound 8 acts as a substrate of Zika virus protease and only the non-prime site residues remain bound to the protease, indicating the possibility of product inhibition (Fig. 4).
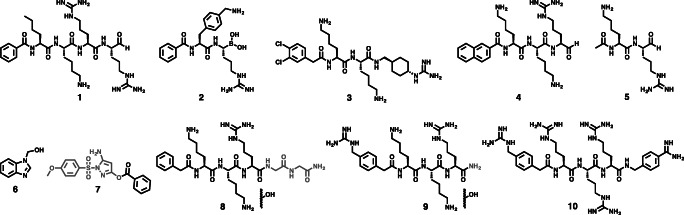
Fig. 4.
Active-site inhibitors that were co-crystallised with NS2B-NS3 proteases from dengue, West Nile and Zika viruses. The actual observed ligand is highlighted in black. Inhibitor moieties that have been cleaved or hydrolysed by proteolytic digest are shown in grey. Compounds 1, 2, 4, 5 and 7 bind covalently to the catalytically active residue S135
Conclusion
Despite two decades of active research, the suitability of flaviviral proteases to serve as drug targets is yet to be proven. For a long time, the debate was dominated by subtle differences and structural uncertainties in artificial model systems that were mainly designed for screening campaigns and only reflected the in vivo situation to a limited degree. This has constrained efforts to identify promising lead compounds and distracted the attention from the actual main challenge: How drug-like and high-affinity compounds can be generated, given the fact of such an unfortunate dibasic substrate recognition motif. Options are available to address this dilemma: First, active-site inhibitors that are structurally unrelated to the substrate should be pursued more intensively. Fragment-based techniques could be one way of doing this and the recently solved first co-crystal structure of a flavivirus protease with an active-site fragment (Zhang et al. 2016) may be a promising step in the right direction. Second, highly modified, metabolically stable and target-selective substrate mimetics might reach the protease in vivo, if modern drug delivery concepts are explored. Third, yet underexplored allosteric inhibitors may offer a convenient option to circumvent the difficulties associated with active-site ligands. The upcoming decade of research will show whether one of these avenues will provide the first clinical candidate to combat diseases associated with dengue, West Nile and Zika virus infections.
Acknowledgments
Christoph Nitsche thanks the Alexander von Humboldt Foundation for a Feodor Lynen Fellowship and the Free University of Berlin for a Rising Star Fellowship.
Compliance with ethical standards
Conflict of interest
Christoph Nitsche declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows NJ, Campos RK, Liao KC, Prasanth KR, Soto-Acosta R, Yeh SC, Schott-Lerner G, Pompon J, Sessions OM, Bradrick SS, Garcia-Blanco MA. Biochemistry and molecular biology of flaviviruses. Chem Rev. 2018;118:4448–4482. doi: 10.1021/acs.chemrev.7b00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. An update on Zika virus infection. Lancet. 2017;390:2099–2109. doi: 10.1016/s0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- Behnam MA, Graf D, Bartenschlager R, Zlotos DP, Klein CD. Discovery of nanomolar dengue and West Nile virus protease inhibitors containing a 4-benzyloxyphenylglycine residue. J Med Chem. 2015;58:9354–9370. doi: 10.1021/acs.jmedchem.5b01441. [DOI] [PubMed] [Google Scholar]
- Behnam MA, Nitsche C, Boldescu V, Klein CD. The medicinal chemistry of dengue virus. J Med Chem. 2016;59:5622–5649. doi: 10.1021/acs.jmedchem.5b01653. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldescu V, Behnam MAM, Vasilakis N, Klein CD. Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov. 2017;16:565–586. doi: 10.1038/nrd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher M, Li Z, Liu B, Zhang J, Koetzner CA, Alifarag A, Jones SA, Lin Q, Kramer LD, Li H. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017;13:e1006411. doi: 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki T. Increase of West Nile virus cases in Europe for 2018. Lancet. 2018;392:1000. doi: 10.1016/S0140-6736(18)32286-4. [DOI] [PubMed] [Google Scholar]
- Chandramouli S, Joseph JS, Daudenarde S, Gatchalian J, Cornillez-Ty C, Kuhn P. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J Virol. 2010;84:3059–3067. doi: 10.1128/jvi.02044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yang K, Wu C, Chen C, Hu C, Buzovetsky O, Wang Z, Ji X, Xiong Y, Yang H. Mechanisms of activation and inhibition of Zika virus NS2B-NS3 protease. Cell Res. 2016;26:1260–1263. doi: 10.1038/cr.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksupmanee O, Hodge K, Katzenmeier G, Chimnaronk S. Structural platform for the autolytic activity of an intact NS2B-NS3 protease complex from dengue virus. Biochemistry. 2012;51:2840–2851. doi: 10.1021/bi2018267. [DOI] [PubMed] [Google Scholar]
- de la Cruz L, Nguyen TH, Ozawa K, Shin J, Graham B, Huber T, Otting G. Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: fold analysis by pseudocontact shifts. J Am Chem Soc. 2011;133:19205–19015. doi: 10.1021/ja208435s. [DOI] [PubMed] [Google Scholar]
- de la Cruz L, Chen WN, Graham B, Otting G. Binding mode of the activity-modulating C-terminal segment of NS2B to NS3 in the dengue virus NS2B-NS3 protease. FEBS J. 2014;281:1517–1533. doi: 10.1111/febs.12729. [DOI] [PubMed] [Google Scholar]
- ECDC (2018) Epidemiological update: West Nile virus transmission season in Europe, 2018. https://ecdc.europa.eu/en/news-events/epidemiological-update-west-nile-virus-transmission-season-europe-2018. Accessed 21 February 2019
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Hammamy MZ, Haase C, Hammami M, Hilgenfeld R, Steinmetzer T. Development and characterization of new peptidomimetic inhibitors of the West Nile virus NS2B-NS3 protease. ChemMedChem. 2013;8:231–241. doi: 10.1002/cmdc.201200497. [DOI] [PubMed] [Google Scholar]
- Hill ME, Yildiz M, Hardy JA (2018) Cysteine disulfide traps reveal distinct conformational ensembles in dengue virus NS2B-NS3 protease. Biochemistry. 10.1021/acs.biochem.8b00978 [DOI] [PubMed]
- Huang Q, Li Q, Joy J, Chen AS, Ruiz-Carrillo D, Hill J, Lescar J, Kang C. Lyso-myristoyl phosphatidylcholine micelles sustain the activity of dengue non-structural (NS) protein 3 protease domain fused with the full-length NS2B. Protein Expr Purif. 2013;92:156–162. doi: 10.1016/j.pep.2013.09.015. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Phillips J, Shun TY, Shinde S, Lazo JS, Huryn DM, Myers MC, Ratnikov BI, Smith JW, Su Y, Dahl R, Cosford ND, Shiryaev SA, Strongin AY. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- Kang C, Keller TH, Luo D. Zika virus protease: an antiviral drug target. Trends Microbiol. 2017;25:797–808. doi: 10.1016/j.tim.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Kim YM, Gayen S, Kang C, Joy J, Huang Q, Chen AS, Wee JL, Ang MJ, Lim HA, Hung AW, Li R, Noble CG, Lee le T, Yip A, Wang QY, Chia CS, Hill J, Shi PY, Keller TH. NMR analysis of a novel enzymatically active unlinked dengue NS2B-NS3 protease complex. J Biol Chem. 2013;288:12891–12900. doi: 10.1074/jbc.M112.442723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh-Stenta X, Joy J, Wang SF, Kwek PZ, Wee JL, Wan KF, Gayen S, Chen AS, Kang C, Lee MA, Poulsen A, Vasudevan SG, Hill J, Nacro K. Identification of covalent active site inhibitors of dengue virus protease. Drug Des Devel Ther. 2015;9:6389–6399. doi: 10.2147/dddt.s94207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Hansen G, Nitsche C, Klein CD, Zhang L, Hilgenfeld R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science. 2016;353:503–505. doi: 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Li J, Lim SP, Beer D, Patel V, Wen D, Tumanut C, Tully DC, Williams JA, Jiricek J, Priestle JP, Harris JL, Vasudevan SG. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J Biol Chem. 2005;280:28766–28774. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- Li Y, Phoo WW, Loh YR, Zhang Z, Ng EY, Wang W, Keller TH, Luo D, Kang C. Structural characterization of the linked NS2B-NS3 protease of Zika virus. FEBS Lett. 2017;591:2338–2347. doi: 10.1002/1873-3468.12741. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Phoo WW, Loh YR, Wang W, Liu S, Chen MW, Hung AW, Keller TH, Luo D, Kang C. Structural dynamics of Zika virus NS2B-NS3 protease binding to dipeptide inhibitors. Structure. 2017;25:1242–1250. doi: 10.1016/j.str.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Z, Phoo WW, Loh YR, Li R, Yang HY, Jansson AE, Hill J, Keller TH, Nacro K, Luo D, Kang C. Structural insights into the inhibition of Zika virus NS2B-NS3 protease by a small-molecule inhibitor. Structure. 2018;26:555–564. doi: 10.1016/j.str.2018.02.005. [DOI] [PubMed] [Google Scholar]
- Lim SP, Shi PY. West Nile virus drug discovery. Viruses. 2013;5:2977–3006. doi: 10.3390/v5122977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SP, Wang QY, Noble CG, Chen YL, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D, Lescar J, Shi PY. Ten years of dengue drug discovery: progress and prospects. Antivir Res. 2013;100:500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/jvi.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, Kotaka M, Lescar J, Vasudevan SG. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahawaththa MC, Pearce BJG, Szabo M, Graham B, Klein CD, Nitsche C, Otting G. Solution conformations of a linked construct of the Zika virus NS2B-NS3 protease. Antivir Res. 2017;142:141–147. doi: 10.1016/j.antiviral.2017.03.011. [DOI] [PubMed] [Google Scholar]
- Mahawaththa MC, Lee MD, Giannoulis A, Adams LA, Feintuch A, Swarbrick JD, Graham B, Nitsche C, Goldfarb D, Otting G. Small neutral Gd(III) tags for distance measurements in proteins by double electron-electron resonance experiments. Phys Chem Chem Phys. 2018;20:23535–23545. doi: 10.1039/c8cp03532f. [DOI] [PubMed] [Google Scholar]
- Nall TA, Chappell KJ, Stoermer MJ, Fang NX, Tyndall JD, Young PR, Fairlie DP. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J Biol Chem. 2004;279:48535–48542. doi: 10.1074/jbc.M406810200. [DOI] [PubMed] [Google Scholar]
- Ng EY, Loh YR, Li Y, Li Q, Kang C. Expression, purification of Zika virus membrane protein-NS2B in detergent micelles for NMR studies. Protein Expr Purif. 2019;154:1–6. doi: 10.1016/j.pep.2018.09.013. [DOI] [PubMed] [Google Scholar]
- Nitsche C. Strategies towards protease inhibitors for emerging flaviviruses. In: Hilgenfeld R, Vasudevan SG, editors. Dengue and Zika: control and antiviral treatment strategies. Singapore: Springer; 2018. pp. 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche C, Holloway S, Schirmeister T, Klein CD. Biochemistry and medicinal chemistry of the dengue virus protease. Chem Rev. 2014;114:11348–11381. doi: 10.1021/cr500233q. [DOI] [PubMed] [Google Scholar]
- Nitsche C, Zhang L, Weigel LF, Schilz J, Graf D, Bartenschlager R, Hilgenfeld R, Klein CD. Peptide–boronic acid inhibitors of flaviviral proteases: medicinal chemistry and structural biology. J Med Chem. 2017;60:511–516. doi: 10.1021/acs.jmedchem.6b01021. [DOI] [PubMed] [Google Scholar]
- Nitsche C, Passioura T, Varava P, Mahawaththa MC, Leuthold MM, Klein CD, Suga H, Otting G (2019) De novo discovery of nonstandard macrocyclic peptides as noncompetitive inhibitors of the Zika virus NS2B-NS3 protease. ACS Med Chem Lett 10:168–174. 10.1021/acsmedchemlett.8b00535 [DOI] [PMC free article] [PubMed]
- Noble CG, Seh CC, Chao AT, Shi PY. Ligand-bound structures of the dengue virus protease reveal the active conformation. J Virol. 2012;86:438–446. doi: 10.1128/jvi.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phoo WW, Li Y, Zhang Z, Lee MY, Loh YR, Tan YB, Ng EY, Lescar J, Kang C, Luo D (2016) Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat Commun 7(13410). 10.1038/ncomms13410 [DOI] [PMC free article] [PubMed]
- Phoo WW, Zhang Z, Wirawan M, Chew EJC, Chew ABL, Kouretova J, Steinmetzer T, Luo D. Structures of Zika virus NS2B-NS3 protease in complex with peptidomimetic inhibitors. Antivir Res. 2018;160:17–24. doi: 10.1016/j.antiviral.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Poland GA, Kennedy RB, Ovsyannikova IG, Palacios R, Ho PL, Kalil J. Development of vaccines against Zika virus. Lancet Infect Dis. 2018;18:e211–e219. doi: 10.1016/s1473-3099(18)30063-x. [DOI] [PubMed] [Google Scholar]
- Poulsen A, Kang C, Keller TH. Drug design for flavivirus proteases: what are we missing? Curr Pharm Des. 2014;20:3422–3427. doi: 10.2174/13816128113199990633. [DOI] [PubMed] [Google Scholar]
- Robin G, Chappell K, Stoermer MJ, Hu SH, Young PR, Fairlie DP, Martin JL. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J Mol Biol. 2009;385:1568–1577. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- Roy A, Lim L, Srivastava S, Lu Y, Song J. Solution conformations of Zika NS2B-NS3pro and its inhibition by natural products from edible plants. PLoS One. 2017;12:e0180632. doi: 10.1371/journal.pone.0180632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- Schüller A, Yin Z, Brian Chia CS, Doan DN, Kim HK, Shang L, Loh TP, Hill J, Vasudevan SG. Tripeptide inhibitors of dengue and West Nile virus NS2B-NS3 protease. Antivir Res. 2011;92:96–101. doi: 10.1016/j.antiviral.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Shiryaev SA, Ratnikov BI, Chekanov AV, Sikora S, Rozanov DV, Godzik A, Wang J, Smith JW, Huang Z, Lindberg I, Samuel MA, Diamond MS, Strongin AY. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393:503–511. doi: 10.1042/bj20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Farhy C, Pinto A, Huang CT, Simonetti N, Elong Ngono A, Dewing A, Shresta S, Pinkerton AB, Cieplak P, Strongin AY, Terskikh AV. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antivir Res. 2017;143:218–229. doi: 10.1016/j.antiviral.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidique S, Shiryaev SA, Ratnikov BI, Herath A, Su Y, Strongin AY, Cosford ND. Structure-activity relationship and improved hydrolytic stability of pyrazole derivatives that are allosteric inhibitors of West Nile virus NS2B-NS3 proteinase. Bioorg Med Chem Lett. 2009;19:5773–5777. doi: 10.1016/j.bmcl.2009.07.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoermer MJ, Chappell KJ, Liebscher S, Jensen CM, Gan CH, Gupta PK, Xu WJ, Young PR, Fairlie DP. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. J Med Chem. 2008;51:5714–5721. doi: 10.1021/jm800503y. [DOI] [PubMed] [Google Scholar]
- Su XC, Ozawa K, Qi R, Vasudevan SG, Lim SP, Otting G. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl Trop Dis. 2009;3:e561. doi: 10.1371/journal.pntd.0000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- Timiri AK, Sinha BN, Jayaprakash V. Progress and prospects on DENV protease inhibitors. Eur J Med Chem. 2016;117:125–143. doi: 10.1016/j.ejmech.2016.04.008. [DOI] [PubMed] [Google Scholar]
- WHO Dengue vaccine: WHO position paper – September 2018. Wkly Epidemiol Rec. 2018;93:457–476. [Google Scholar]
- Wu H, Bock S, Snitko M, Berger T, Weidner T, Holloway S, Kanitz M, Diederich WE, Steuber H, Walter C, Hofmann D, Weissbrich B, Spannaus R, Acosta EG, Bartenschlager R, Engels B, Schirmeister T, Bodem J. Novel dengue virus NS2B/NS3 protease inhibitors. Antimicrob Agents Chemother. 2015;59:1100–1109. doi: 10.1128/aac.03543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz M, Ghosh S, Bell JA, Sherman W, Hardy JA. Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem Biol. 2013;8:2744–2752. doi: 10.1021/cb400612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Patel SJ, Wang W-L, Wang G, Chan W-L, Rao KRR, Alam J, Jeyaraj DA, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Peptide inhibitors of dengue virus NS3 protease. Part 1: warhead. Bioorg Med Chem Lett. 2006;16:36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Y, Loh YR, Phoo WW, Hung AW, Kang C, Luo D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science. 2016;354:1597–1600. doi: 10.1126/science.aai9309. [DOI] [PubMed] [Google Scholar]