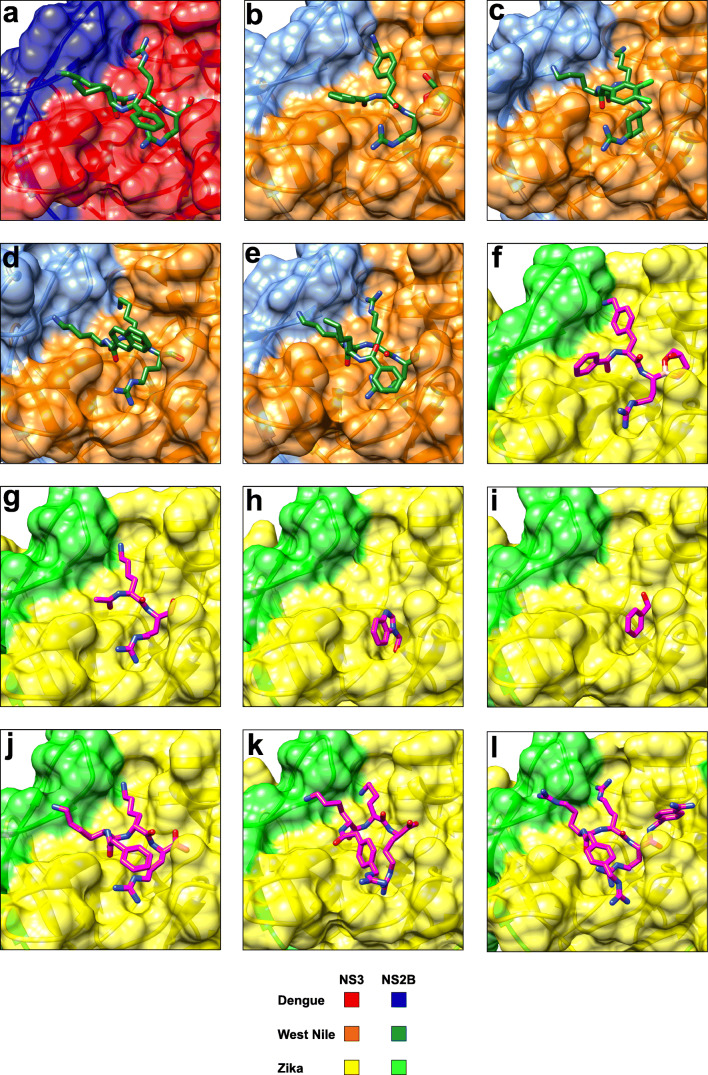
Fig. 3.
X-ray co-crystal structures of flavivirus NS2B-NS3 proteases in complex with active-site inhibitors. (a) Dengue protease serotype 3 in complex with compound 1 (3U1I) (Noble et al. 2012). The aldehyde in 1 forms a covalent hemiacetal with S135 (not shown). (b) West Nile protease in complex with compound 2 (5IDK) (Nitsche et al. 2017). The boronic acid in 2 forms a cyclic ester adduct with glycerol (as shown) and a covalent boronate with S135 (not shown). (c) West Nile protease in complex with compound 3 (2YOL) (Hammamy et al. 2013). (d) West Nile protease in complex with compound 4 (3E90) (Robin et al. 2009). The aldehyde in 4 forms a covalent hemiacetal with S135 (not shown). (e) West Nile protease in complex with compound 1 (2FP7) (Erbel et al. 2006). The aldehyde in 1 forms a covalent hemiacetal with S135 (not shown). (f) Zika protease in complex with compound 2 (5LC0) (Lei et al. 2016). The boronic acid in 2 forms a cyclic ester adduct with glycerol (as shown) and a covalent boronate with S135 (not shown). (g) Zika protease C143S mutant in complex with compound 5 (5YOF) (Li et al. 2018). The aldehyde in 5 forms a covalent hemiacetal with S135 (not shown). A similar structure of lower resolution has also been reported for compound 5 in complex with the Zika protease wildtype (5H6V) (Li et al. 2017b). (h) Zika protease in complex with fragment 6 (5H4I) (Zhang et al. 2016). (i) Zika protease in complex with a benzoyl fragment (5YOD) (Li et al. 2018). Transesterification between Zika protease and compound 7 results in the formation of a S135 benzoate (covalent bond not shown). (j) Zika protease C143S mutant in complex with proteolytically cleaved compound 8 (5ZMQ) (Phoo et al. 2018). (k) Zika protease C143S mutant in complex with proteolytically hydrolysed compound 9 (5ZMS) (Phoo et al. 2018). (l) Zika protease C143S mutant in complex with compound 10 (5ZOB) (Phoo et al. 2018). This figure has been generated with Chimera (Pettersen et al. 2004)