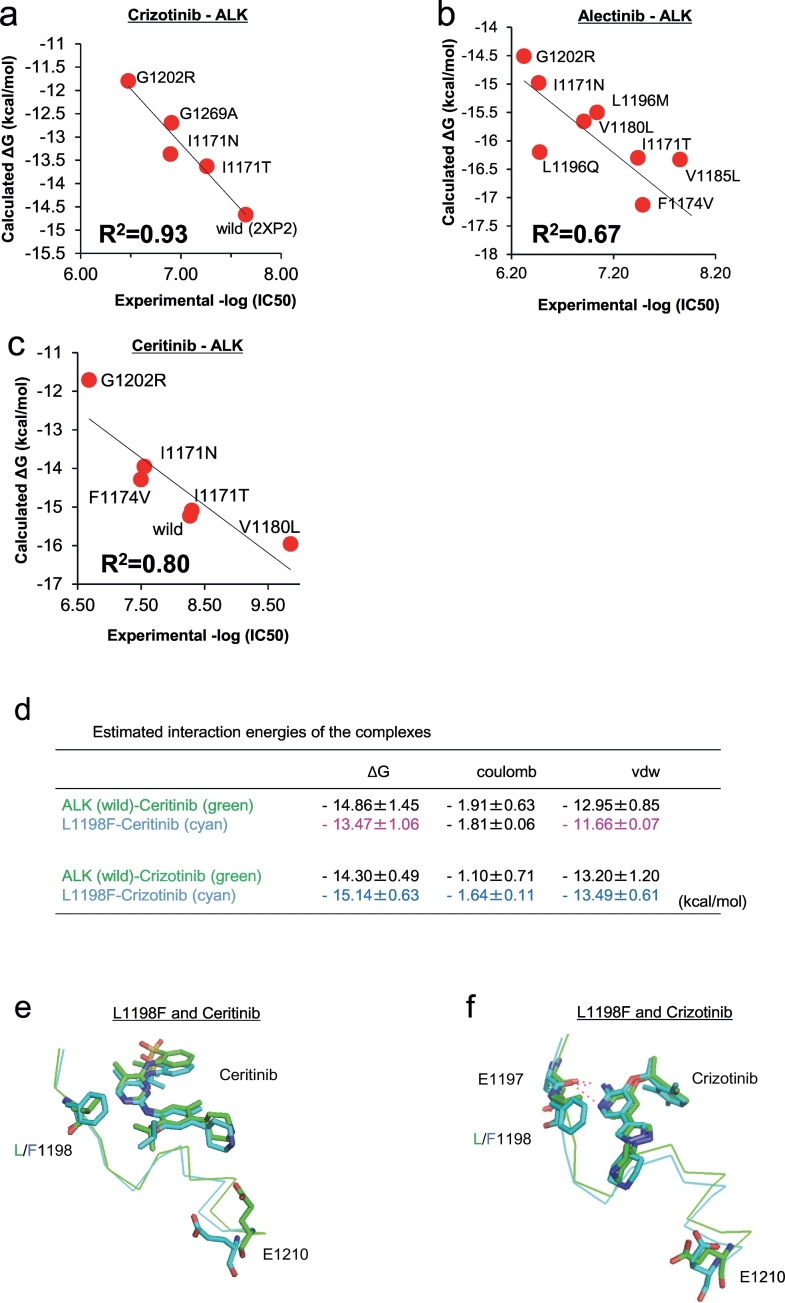
Fig. 4.
Computational prediction of the binding affinity between ALK mutants and ALK-TKIs.
(a–c) The binding free energy (ΔG) of crizotinib (a), alectinib (b), or ceritinib (c) to WT or each resistant mutant is plotted against experimental IC50 of the corresponding Ba/F3 mutant. These ΔG values are calculated by MP-CAFEE, which is one of the MD free energy simulation methods. Solid lines represent linear fits with square of the correlation coefficients (R2) of 0.93, 0.67, and 0.80 for crizotinib-ALK, alectinib-ALK, and ceritinib-ALK plots, respectively. (d) The average binding free energies (ΔG values) of ceritinib or crizotinib (from three sets of free energy simulations). Electrostatic (coulomb) and van der Waals (vdw) contributions in ΔG values indicate that this mutation induces a slight increase in electrostatic interactions with crizotinib but decreases the van der Waals contacts with ceritinib. (e and f) Conformational differences in the drug-binding site between ALK-WT (green) and L1198F mutant (cyan). E1197, L/F1198, E1210, and ceritinib (e) or crizotinib (f) in an energetically stable conformation obtained from five sets of 50 ns simulations are depicted by sticks (green/cyan, carbon; blue, nitrogen; red, oxygen; yellow, sulfur). ALK-TKI hydrogen bonds are depicted by dashed lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)