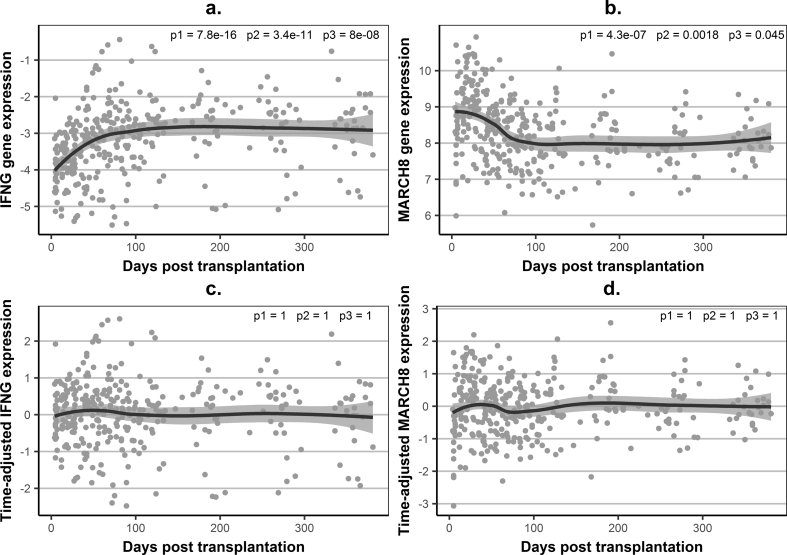
Fig. 2.
Examples of association between gene-expression levels and time in stable patients.
(a) – observed (unadjusted) –ΔCt values for IFNG, as an example of a gene with increasing expression levels; (b) – observed (unadjusted) –ΔCt values for MARCH8, as an example of a gene with decreasing expression levels; (c) – time-adjusted gene-expression levels for IFNG, illustrating abolition of time-dependence for gene expression; (d) – time-adjusted gene-expression levels for MARCH8; Gene expression – observed ΔCt values, relative to hypoxanthine phosphoribosyltransferase (HPRT), in individual samples (grey dots) and a loess average curve with 95% confidence interval; Time-adjusted gene expression – fold difference on log2 scale (same scale as gene expression), derived as the difference between the observed –ΔCt values and the predicted values using generalised linear mixed-effects models (GLMM) linear regression, based on all serial samples (total n = 335) from the training stable kidney transplant recipients (n = 27) (a cubic relationship between gene expression and time post transplantation was modelled for the fixed and random effects); p1, p2, p3 - p-values for the linear, quadratic and cubic fixed-effects terms for time in GLMM (in (a) and (b) GLMM was based on the observed –ΔCt values and the p-values confirm a statistically significant association between gene-expression and time, in (c) and (d) GLMM was based on time-adjusted gene expression values and p-values demonstrate the time-independence of the time-adjusted gene-expression levels).