Fig. 1.
Overview of patient samples and data sets used for integrated analyses.
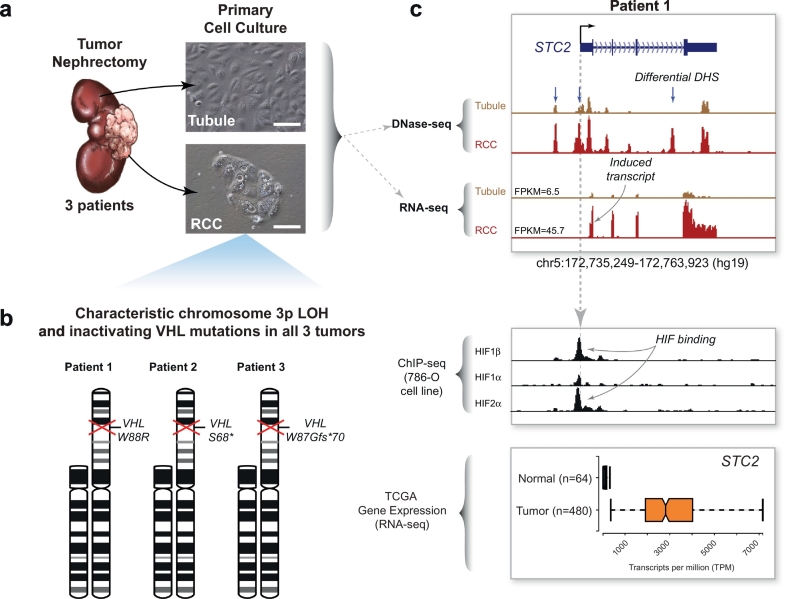
(a) Primary culture of tumor and matched-normal tubule cells from three patients. Renal cell carcinoma tumor nephrectomies from three patients were used to derive primary cultures of proximal tubules and renal cell carcinoma. Microscope images scale bar = 100 μm.
(b) Cytogenetic analysis of primary tumor cultures. Karyotype analysis of the carcinoma cultures revealed loss of the short of arm of chromosome 3 in all three patient samples. Sanger sequencing of the VHL gene in these same samples identified inactivating mutations in the remaining copy.
(c) Example of integrated analysis at the STC2 gene locus. DNase-seq and RNA-seq datasets were also generated from the primary tubule and carcinoma cultures and compared to HIF ChIP-seq datasets from the 786-O renal cell carcinoma cell line and RNA-seq expression data from TCGA. STC2, a canonical HIF target gene, exhibited several differential DHS (blue arrows), some of which coincided with HIF binding determined by ChIP-seq. Compared to normal tubules, the STC2 transcript was strongly induced in the primary tumor cultures and in the TCGA tumor samples (depicted with 10% outlier trim for clarity).