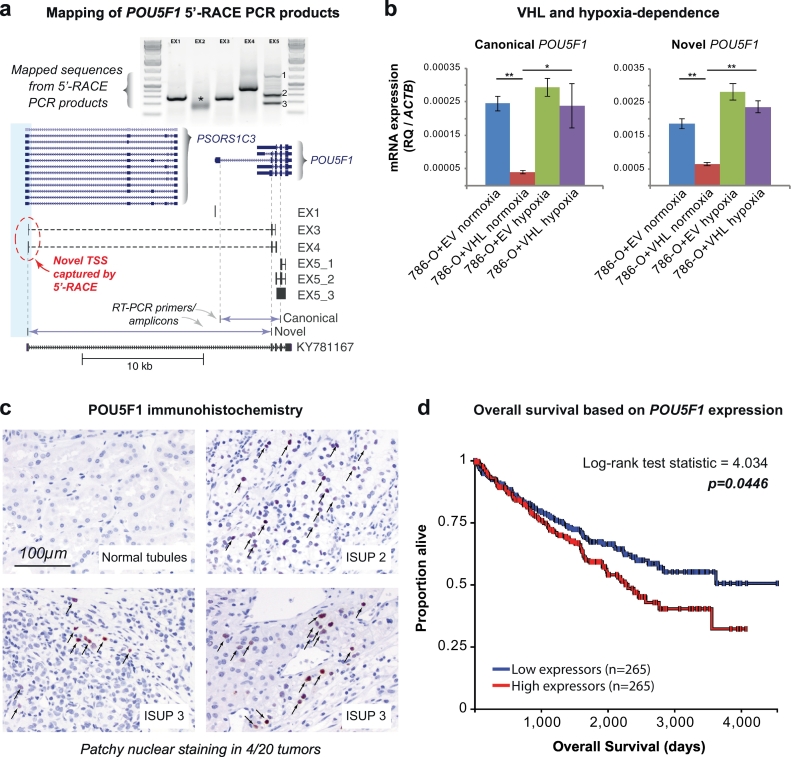
Fig. 6.
The novel transcript for POU5F1 exhibits HIF dependence and POU5F1 expression levels correlate with patient survival.
(a) Schematic mapping of POU5F1 5′-RACE PCR products. 5′-RACE performed on 786-O RNA captured a transcription start site originating in the -16 kb DHS (blue shaded box) therefore defining a novel isoform of POU5F1 that is produced following read through of the PSORS1C3 gene. Reverse primers in known POU5F1 exons (e.g. EX1 = reverse primer in exon 1) were used to amplify the 5′-ends of the cDNA molecule captured by 5′-RACE and sequence mapped to the genome. The exon 2 primer (*) failed to yield mappable sequence. The exon 5 primer yielded 3 different products (EX5-1, EX5-2, EX5-3). The location of RT-PCR primers to detect the canonical and novel POU5F1 transcript variants are also indicated. The inferred isoform structure is compared to EST KY781167.
(b) Canonical and novel POU5F1 transcripts exhibit HIF-dependence. RT-PCR primers were used to quantify the canonical and novel POU5F1 transcripts in 786-O cells stably transduced with VHL (786-O + VHL) or empty vector (786-O + EV) cultured in normoxia or hypoxia (2% O2) for 24 h. Expression levels (relative quantification, RQ) were calculated using the β-actin housekeeping gene (ACTB). Compared to the empty vector control, reintroduction of VHL protein into 786-O cells suppressed expression of both POU5F1 transcripts. Exposing 786-O + VHL cells to hypoxia induced both POU5F1 transcripts. N.B. expression scale differences between the canonical and novel transcripts. Error bars indicate standard deviations of three experimental replicates. *p < 0.05, **p < 0.005.
(c) Immunohistochemistry of POU5F1 protein in renal cell carcinoma samples. POU5F1 (OCT4) immunohistochemistry was performed on RCC samples from 20 patients (5 from each of ISUP grades 1–4) and showed patchy nuclear positivity (arrows) in a single random sample from 4 patients. No nuclear staining was seen in any of the matched normal renal parenchyma from the same patients.
(d) Overall survival as a function of POU5F1 expression in TCGA. Patients with POU5F1 expression data from TCGA (KIRC) were evenly divided into two groups split at the median expression level (233 RSEM normalized) and Kaplan-Meier curves for overall survival were plotted using the UCSC Xena browser tool.