Figure 4.
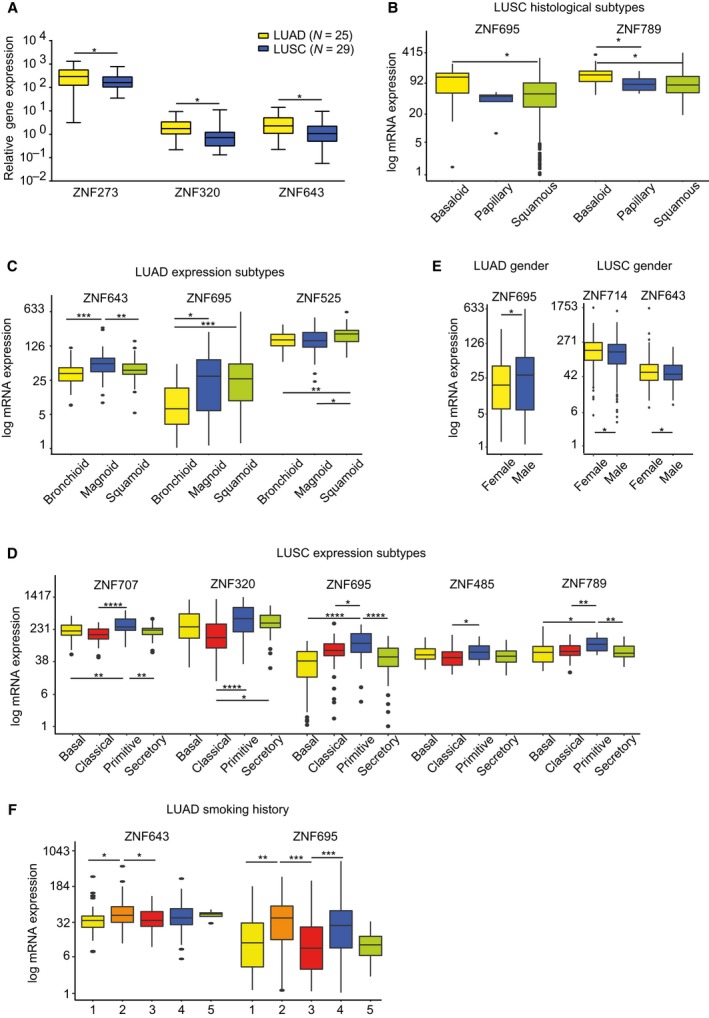
Cancer‐associated KRAB‐ZNFs correlate with various clinical parameters in LUAD and LUSC. (A) Single‐gene qPCR analysis in lung cancer tissue panel shows that ZNF273,ZNF320, and ZNF643 have significantly higher mRNA level in LUAD (n = 25) compared to LUSC (n = 29). (B) In TCGA LUSC histological subtypes, ZNF695 and ZNF789 demonstrate higher expression in the basaloid (n = 15) subtype compared to other subtypes: papillary (n = 6) and/or squamous not otherwise specified (n = 467). (C) Boxplot representation of mRNA expression level of ZNF525,ZNF643, and ZNF695 in TCGA LUAD molecular subtypes: squamoid (n = 78), bronchoid (n = 89), and magnoid (n = 63). (D) Boxplot representation of mRNA expression level of ZNF707,ZNF320,ZNF695,ZNF485, and ZNF789 in TCGA LUSC molecular subtypes: basal (n = 42), classical (n = 65), primitive (n = 27), and secretory (n = 43). (E) The expression of ZNF695 in TCGA LUAD, as well as ZNF714 and ZNF643 in TCGA LUSC correlate with gender. LUAD: female (n = 277), male (n = 239); LUSC: female (n = 130), male (n = 370). (F) ZNF643 and ZNF695 mRNA is more abundant in smokers vs nonsmokers or reformed smokers according to the TCGA dataset. 1: lifelong nonsmoker (n = 76), 2: current smoker (n = 119), 3: current reformed smoker for > 15 years (n = 135), 4: current reformed smoker for ≤ 15 years (n = 168), 5: current reformed smoker, duration not specified (n = 4). (A–F) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as assessed by t‐test with FDR correction.
