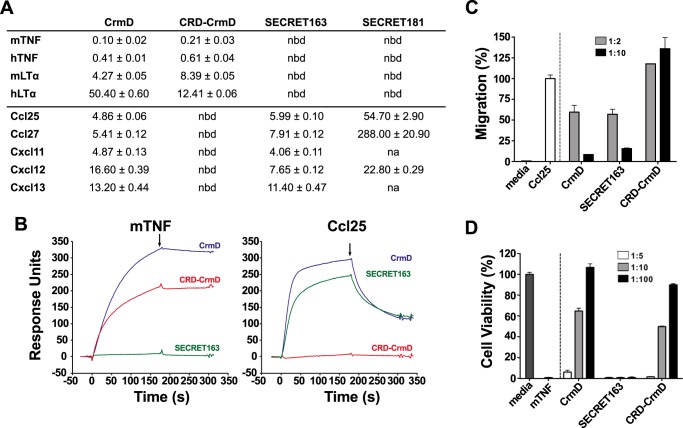
Figure 2.
SECRET163, but not SECRET181, displays chemokine-binding and -inhibitory activities comparable with those of full-length CrmD. A, TNFSF ligand- and chemokine-binding affinity constants (KD) of the CrmD truncated mutants calculated by SPR. The table shows the mean ± S.D. KD in nm units. nbd, no binding detected; na, not assayed. B, SPR sensorgrams showing the binding of 100 nm mTNF or mouse Ccl25 to CM4 chips immobilized with CrmD (blue), CRD-CrmD (red), or SECRET163 (green). Black arrows indicate the end of the analyte injection. C, MOLT-4 cell chemotaxis assay. MOLT-4 transwell migration was induced by 70 nm mouse Ccl25 preincubated in the absence (Ccl25) or presence of recombinant protein at the indicated chemokine:protein molar ratios (see legend) of CrmD, SECRET163, or CRD-CrmD. Migrated cells were detected using a Cell Titer Aqueous One Solution kit and measuring the A490 in a microplate reader. Media indicates the cell migration detected in the absence of chemokines. Data are represented as the percentage of migration relative to the A490 detected when the cells were incubated with the chemokine alone (Ccl25). D, mTNF-induced cytotoxicity assay on L929 cells. L929 cells were incubated with 1.2 nm mTNF in the absence or presence of recombinant protein at increasing mTNF:protein molar ratios (see legend). Cell viability was assessed as the A490 determined using a Cell Titer Aqueous One Solution kit. The A490 calculated for cells incubated with mTNF alone (mTNF) was subtracted from all samples. Data are represented as the percentage relative to the A490 recorded for cells incubated without mTNF (media). In C and D, results are shown as mean ± S.D. (error bars) of triplicates of one experiment representative of three independent experiments.