Abstract
Isoprenyl cysteine carboxyl methyltransferase (ICMT) plays a key role in post-translational regulation of prenylated proteins. On the basis of previous results, we hypothesized that the p53 pathway and ICMT expression may be linked in cancer cells. Here, we studied whether WT p53 and cancer-associated p53 point mutants regulate ICMT levels and whether ICMT overexpression affects tumor progression. Studying the effect of p53 variants on ICMT mRNA and protein levels in cancer cells, we found that WT p53 and p53 mutants differentially affect ICMT expression, indicating that p53 status influences ICMT levels in tumors. To investigate the underlying mechanisms, we constructed ICMT–luciferase reporters and found that WT p53 represses ICMT transcription. In contrast, p53 mutants showed a positive effect on ICMT expression. Promoter truncation analyses pinpointed the repressive effect of WT p53 to the −209 and −14 region on the ICMT promoter, and ChIP assays indicated that WT p53 is recruited to this region. Instead, a different promoter region was identified as responsible for the mutant p53 effect. Studying the effect of ICMT overexpression on tumor-associated phenotypes in vitro and in vivo, and analyzing breast and lung cancer databases, we identified a correlation between p53 status and ICMT expression in breast and lung cancers. Moreover, we observed that ICMT overexpression is correlated with negative clinical outcomes. Our work unveils a link between postprenylation protein processing and the p53 pathway, indicating that the functional interplay between WT and mutant p53 alters ICMT levels, thereby affecting tumor aggressiveness.
Keywords: protein isoprenylation, protein carboxylation, p53, cytoskeleton, breast cancer, lung cancer, cell signaling, gene regulation, post-translational modification (PTM), mutant p53
Introduction
ICMT2 is an enzyme involved in a complex post-translational modification process. The initial step involves the addition of farnesyl or geranylgeranyl isoprenoid to a cysteine residue close to the C terminus of protein substrates (1), catalyzed by farnesyl transferase or geranylgeranyl transferase I. The prenylated cysteine residue is usually part of a CAAX motif (C is cysteine; A is aliphatic amino acid; and X is any amino acid), but other motifs such as CXC can also be targeted by prenyltransferases (2). Then, the terminal amino acids following the prenylated cysteine are eliminated by a specific peptidase known as RCE1, at the endoplasmic reticulum (3). Finally, ICMT catalyzes methylation of the free carboxyl end on cysteine. This modification provides a hydrophobic uncharged C terminus to the substrate protein that increases the interaction with biological membranes and/or modifies the ability to interact with protein partners (4). The existence of more than 200 CAAX proteins has been predicted based on sequence and structural analyses (5, 6). An intriguing aspect of prenylated proteins is that they are distributed among different families and are involved in a variety of biological functions (7).
The identification of RAS family members as ICMT substrates reinforced the notion that protein prenylation may play a role in cancer (8, 9), as suggested by pioneering reports on the inhibitory effect of 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors on cell proliferation (10–12). Supporting this hypothesis, genetic ablation of ICMT reduced KRAS-induced transformation of mouse embryo fibroblasts in vitro (13). Tissue-specific deletion of ICMT in mice expressing mutant KRAS in myeloid cells and pneumocytes, attenuated myeloproliferative syndrome, and reduced the areas of neoplastic lesions in the lungs (14). Supporting a role in RAS-driven tumorigenesis, genetic ablation of ICMT in breast cancer cell lines harboring mutant RAS, reduced tumor formation in a xenograft model (15). In contrast, ICMT can also cooperate with tumor suppression, because its inactivation in pancreatic progenitor cells increased the number of KRAS-driven intraepithelial neoplasias and promoted the progression to pancreatic ductal adenocarcinoma (16). Some RHO GTPase family members are also modified by ICMT (17), providing other potential connections with oncogenic mechanisms. For example, the involvement of RHO family members on actin cytoskeleton remodeling and cell motility suggests that they may affect invasiveness and metastasis (18). ICMT inhibition reduced migration and invasion in MDA-MB-231 cells in vitro (19), concomitant with decreased RHOA and RAC1 activity. Likewise, a decrease in migration and in vivo invasion associated with RAB4A-impaired function was observed in HT-1080 fibrosarcoma cells upon ICMT inhibition (20). Nevertheless, a complex scenario may be envisaged, because the initial view proposing that RHO GTPases play a pro-neoplastic role has been challenged by recent data from in vivo models and human tumors (21). Furthermore, ICMT deregulation is expected to exert complex effects on RHO GTPases because the action of ICMT on specific substrates may have different and even opposing consequences on subcellular localization and/or expression levels (22–24). This evidence indicates that ICMT cannot be considered a proto-oncogene under all circumstances and that several aspects of its biological role are still underexplored. In this context, characterizing the connection between ICMT function and specific oncogenic signaling circuits may help to understand its contribution to cancer.
Despite the growing interest on ICMT, little attention has been paid to the mechanisms that control its expression. Previous results from microarray analysis (25) suggested to us that ICMT expression may be enhanced by mutant p53. The role of p53 as a tumor suppressor has been extensively documented by a large body of evidence, showing that it constitutes the central hub of a signaling pathway activated in response to stress signals (26). Accordingly, the presence of mutations in the p53 gene (TP53) is the most frequent genetic alteration in human cancer (27, 28), characterized by the conspicuous expression of point mutants that may acquire neomorphic activities (29). The presence of mutant p53 proteins may have different consequences on cell physiology (30). On the one hand, p53 point mutants may inhibit WT p53; on the other hand, they may also activate WT p53-independent mechanisms, through new capabilities conferred by the mutation. Compelling evidence from mouse models showed that the expression of p53 point mutants promotes the development of aggressive tumors (31). Nevertheless, the rationalization of the functional complexity of p53 mutants as well as their precise role in different cancer types remains a challenge.
To explore the existence of a link between the p53 pathway and post-prenylation processing, we studied the effect of WT and p53 mutant forms on ICMT expression. We also analyzed the consequences of altering ICMT levels on tumor phenotypes, and we studied the impact of ICMT expression in breast and lung cancer.
Results
Mutant and WT p53 exert opposing effects on ICMT expression
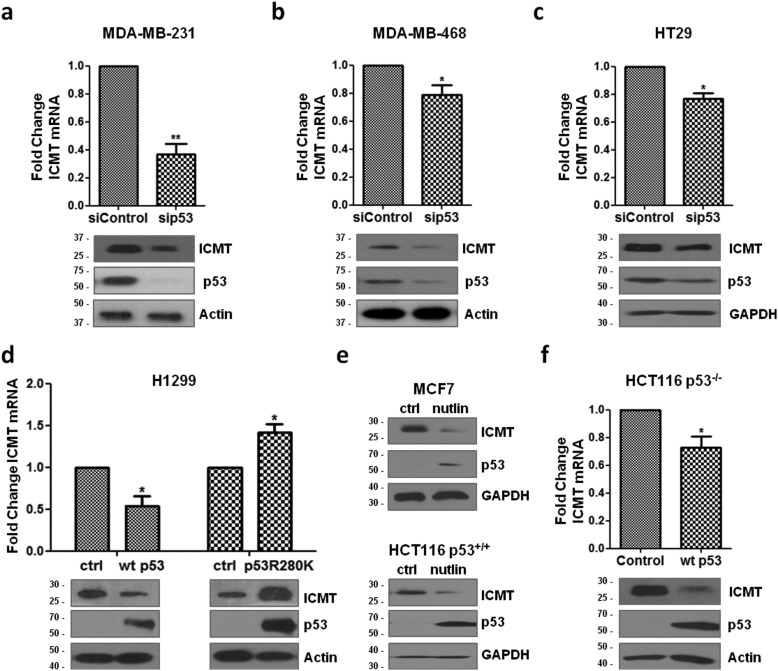
The presence of p53 point mutants was proposed to affect gene expression in tumor cells through different mechanisms. Previous results from microarray analysis on MDA-MB-231 breast cancer cells (25), which endogenously express p53R280K and lack a functional WT TP53 allele, indicated to us that ICMT expression was reduced upon mutant p53 knockdown, thus suggesting a positive regulation. To test this hypothesis, we first confirmed the effect of endogenous p53R280K knockdown on ICMT mRNA steady-state levels in independent experiments. Upon siRNA transfection in MDA-MB-231 cells, total RNA was extracted, and qPCR was performed on cDNA samples. We found that knockdown of endogenous mutant p53 reduced ICMT mRNA levels (Fig. 1a). Accordingly, Western blot analysis showed that ICMT protein levels were reduced upon p53R280K knockdown in the same cell line. We performed a similar analysis on MDA-MB-231 cells where p53R280K expression was stably knocked down. Cells were transduced with retroviral plasmids expressing shRNAs that target two different regions on the p53 mRNA: the same region targeted by the p53 siRNA and a second region present in the 3′ UTR (25). Our results confirmed that ICMT mRNA and protein levels were reduced upon p53R280K silencing with both shRNAs (Fig. S1a), thus indicating that the observed changes are not due to off-target effects. To further confirm our results, we analyzed data from an independent GEO dataset, previously published by other authors (32). We found that ICMT expression was significantly reduced in MDA-MB-231 cells when p53R280K was knocked down, as expected (Fig. S1b). To understand whether other point mutants behave similarly, we concentrated on p53R273H, which is one of the most frequent p53 mutants in human cancer (33). A similar effect on ICMT mRNA and protein levels was observed when we knocked down endogenous p53R273H in breast adenocarcinoma MDA-MB-468 and colorectal adenocarcinoma HT29 cells (Fig. 1, b and c). These results are in agreement with the hypothesis that mutant p53 promotes ICMT expression.
Figure 1.
Mutant and WT p53 differentially regulate ICMT expression. a, MDA-MB-231 cells were transfected with p53 or control siRNA (sip53 or siControl), and mRNA levels were determined by qRT-PCR. ICMT mRNA levels were normalized to GAPDH mRNA and expressed as fold change compared with control condition (upper panel, n = 3, one-tailed t test, p = 0.0062). ICMT and p53R280K levels were determined by Western blotting (lower panel). The effect of endogenous p53R273H knockdown on ICMT expression was analyzed similarly in b, MDA-MB-468 cells (n = 3, one-tailed t test, p = 0.048) or c, HT29 cells (n = 3, one-tailed t test, p = 0.0151). d, H1299 cells were transfected with plasmids expressing WT p53, p53R280K, or empty vector (ctrl) as indicated. ICMT mRNA levels were determined by qRT-PCR, normalized relative to GAPDH mRNA, and expressed as fold change compared with control cells (n = 4, one-tailed t test, p = 0.0147 and p = 0.0123, respectively). Western blot analysis of ICMT and p53 protein levels (lower panel). e, MCF-7 (upper panel) or HCT116 p53+/+ cells (lower panel) were treated with nutlin-3, and protein levels were analyzed by Western blotting. f; effect of WT p53 on ICMT mRNA levels was determined by qRT-PCR upon transfection of HCT116 p53−/− cells with a plasmid expressing WT p53 or empty vector as a control (upper panel; n = 4, one-tailed t test, p = 0.0218). ICMT and WT p53 expression was analyzed by Western blotting (lower panel). *, p < 0.05; **, p < 0.01.
Taking into consideration the close functional interplay between mutant and WT p53 forms (30, 34), we decided to study the effect of WT p53 on ICMT expression. To understand whether WT p53 may also affect ICMT mRNA steady-state levels, we transfected H1299 p53 null cells with a plasmid expressing WT p53 and performed qRT-PCR. Expression of WT p53 significantly reduced ICMT mRNA levels compared with control cells transfected with empty vector (Fig. 1d). In addition, ICMT protein levels were reduced upon WT p53 transfection. Conversely, ICMT mRNA and protein levels increased upon expression of p53R280K (Fig. 1d). A similar effect on ICMT protein levels was observed when we expressed other p53 point mutants (R273H or R175H) in H1299 cells (Fig. S1c). These results confirmed that other point mutants affect ICMT expression.
To further explore the regulation of ICMT expression, we analyzed the effect of endogenous WT p53 stabilization on ICMT levels using MCF-7 breast cancer cell line. Cells were treated with nutlin-3, a specific inhibitor of the interaction between p53 and HDM2 ubiquitin ligase (35), and protein levels were analyzed by Western blotting. As expected, WT p53 levels were increased upon treatment, and concomitantly, a reduction of ICMT levels was observed (Fig. 1e). Similar results were obtained when we treated HCT116 p53+/+ colon carcinoma cells. Moreover, reconstitution of WT p53 expression in syngeneic HCT116 p53−/− cells significantly reduced ICMT mRNA and protein levels (Fig. 1f). These results confirmed the opposing effect of both p53 forms on ICMT levels.
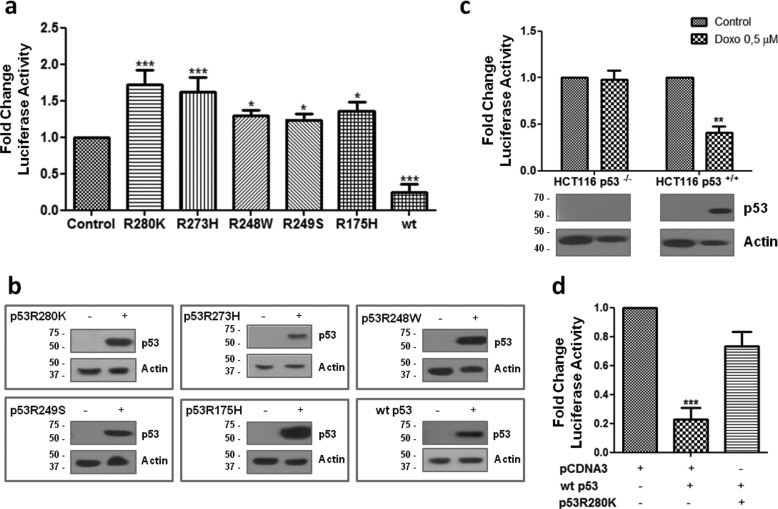
To understand whether p53 forms affect ICMT transcription, we amplified a fragment of the ICMT promoter ranging from −2234 to +37 on MDA-MB-231 genomic DNA, and we cloned it into pGL3-basic vector, to generate a reporter (pICMTluc). Luciferase assays were performed upon co-transfection of a plasmid expressing p53R280K with pICMTluc into H1299 p53 null cells. We found that p53R280K expression significantly enhanced reporter activity in the absence of WT p53 (Fig. 2, a and b). These results are in agreement with our previous observations and support the idea that mutant p53 cooperates with ICMT expression through the acquisition of WT p53-independent activities. Similar experiments were performed co-transfecting pICMTluc with plasmids expressing other p53 point mutants (R273H, R175H, R248W, and R249S). We observed a significant enhancement of reporter activity in all cases (Fig. 2, a and b). In contrast, when we performed luciferase assays in H1299 cells, introducing the WT protein, we found that the activity of the promoter was markedly repressed (Fig. 2, a and b), indicating that ICMT transcription is negatively regulated by the p53 pathway. We also analyzed whether WT p53 induction by doxorubicin treatment may affect ICMT promoter activity. HCT116 p53+/+ or HCT116 p53−/− cells were transfected with pICMTluc and pCMV-β-gal as transfection control, and upon 24 h of treatment, luciferase activity was determined. We found that reporter activity was significantly reduced upon induction of WT p53 in HCT116 p53+/+ cells but was unaffected in HCT116 p53−/− cells (Fig. 2c). These results showed that WT p53 was required for ICMT promoter repression upon induction.
Figure 2.
Mutant and WT p53 differentially regulate the ICMT promoter. a, luciferase assays in H1299 cells co-transfected with pICMTluc, plasmids expressing the indicated proteins or empty vector (control), and pCMV-β-gal. Luciferase values were normalized to β-gal activity and expressed as fold change relative to cells transfected with empty vector (one-tailed t test, *, p < 0.05, and ***, p < 0.0001). b, Western blot analysis confirming p53R280K, p53R273H, p53R248W, p53R249S, p53R175H, and WT p53 expression upon transfection in H1299 cells in luciferase assays from a. c, luciferase assays in HCT116 p53−/− or HCT116 p53+/+ cells co-transfected with pICMTluc and pCMV-β-gal and treated with doxorubicin (upper panel; one-tailed t test, p = 0.0062, **, p < 0.01). Western blot analysis confirming WT p53 induction in HCT116 p53+/+ cells upon doxorubicin treatment (lower panel). d, mutant p53R280K counteracts the repressive effect of WT p53 on the ICMT reporter. Luciferase assays were performed on H1299 cells co-transfected with pICMTluc and plasmids expressing WT p53 and p53R280K as indicated (one-way ANOVA, p < 0.0001).
Considering that WT and mutant p53 forms are expected to co-exist, at least transiently, during tumor progression, we wondered whether the interplay between both p53 forms affects ICMT expression. We co-transfected p53R280K and WT p53 with pICMTluc in H1299 cells and measured reporter activity. We found that the presence of p53R280K almost completely counteracted promoter repression by WT p53 (Fig. 2d). These results suggest that the acquisition of a missense mutation on TP53 would exert a profound effect on ICMT expression by inactivating one WT allele but also because of the dominant-negative effect on the remaining WT protein.
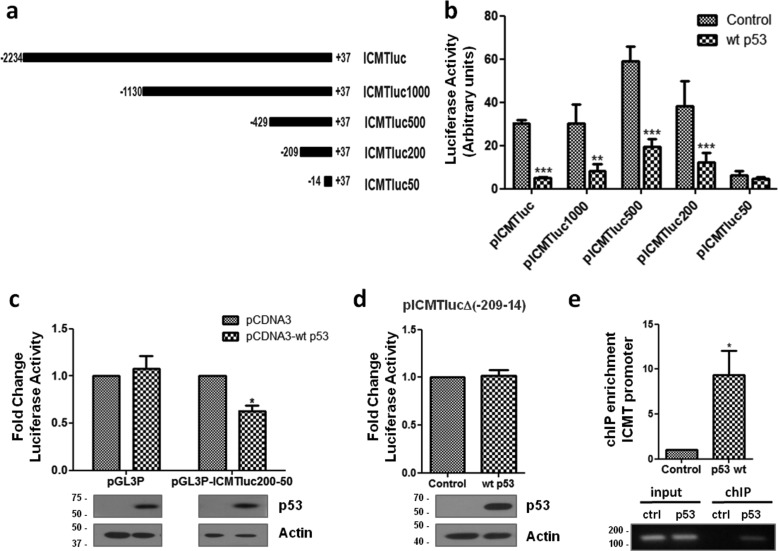
To identify the region responsible for the WT p53-repressive effect, we generated different reporters containing deletions of the ICMT promoter (Fig. 3a). Luciferase assays were performed co-transfecting each reporter with a plasmid expressing WT p53. We found that the repressive effect was maintained on the fragment between positions −209 and +37, but it was almost completely absent on the fragment ranging from −14 to +37 (Fig. 3b), indicating that the region between positions −209 and −14 is required for repression by WT p53. Accordingly, when we cloned the −209 and −14 fragment into pGL3-promoter vector, upstream of the SV40 promoter that drives luciferase transcription, we found that the activity was significantly reduced, compared with control cells (Fig. 3c). Instead, WT p53 showed no effect on empty pGL3-promoter vector, demonstrating that the identified promoter fragment is able to drive WT p53-dependent transcriptional repression.
Figure 3.
Effect of WT p53 on the ICMT promoter. a, schematic representation of ICMT promoter deletions used to generate reporter plasmids. b, luciferase assays on promoter deletions. H1299 cells were co-transfected with the indicated reporters and a plasmid expressing WT p53 or empty vector (control). Luciferase activity was normalized to β-gal and expressed as arbitrary units (two-way ANOVA, **, p < 0.01, and ***, p < 0.001). c, luciferase assays on H1299 cells co-transfected with pCDNA3-WTp53 and pGL3 promoter vector (pGL3P) or pGL3 promoter containing the 195-bp (−209 and −14) fragment (pGL3P-ICMTluc200–50) as indicated. Luciferase activity was normalized and expressed as fold change compared with control cells (transfected with pCDNA3), (one-tailed t test, p = 0.01, *, p < 0.05). Lower panel, confirmation of WT p53 expression by Western blotting. d, luciferase assays on H1299 cells co-transfected with pICMTlucΔ(−209−14) and a plasmid expressing WT p53 or empty vector (control). Luciferase activity was normalized and expressed as fold change compared with control cells. Lower panel, confirmation of WT p53 expression by Western blotting. e, ChIP assay on H1299 cells co-transfected with pICMTluc and pCDNA3-WTp53 or pCDNA3 as a control. IP was performed using anti-p53 antibody (DO1). The presence of ICMT promoter DNA in immunoprecipitates was determined by q-PCR (upper panel, n = 3, one-tailed t test, p = 0.026, *, p < 0.05) and semiquantitative PCR (lower panel).
To confirm that this region is responsible for the WT p53-repressive effect in the context of the ICMT promoter, we generated a reporter containing the −2234 + 37 promoter fragment bearing a deletion between positions −209 and −14 (pICMTlucΔ(−209−14)). As expected, our results showed that the absence of the identified region completely abolished promoter repression by WT p53 (Fig. 3d). To understand whether WT p53 may act directly on the ICMT promoter, we performed ChIP experiments. H1299 cells were transfected with a plasmid expressing WT p53 and pICMTluc, and the protein was immunoprecipitated using an anti-p53 antibody. We found that a 102-bp fragment between positions −66 and +36 on the ICMT promoter could be amplified on the immunoprecipitated DNA from cells expressing WT p53 (Fig. 3e), but not from cells lacking WT p53, confirming the recruitment of the protein on the ICMT promoter.
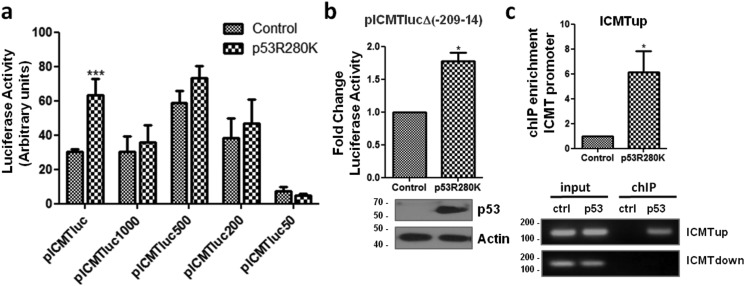
Next, we mapped the region involved in the mutant p53 effect. We performed luciferase assays with the same reporters used for WT p53. A clear reduction on promoter activation by p53R280K was found using all the subsequent reporter deletions, compared with the −2234 + 37 promoter fragment, indicating that the effect of mutant p53 depends on the region between −2234 and −1130 (Fig. 4a). Moreover, p53R280K was able to enhance promoter activity also on pICMTlucΔ(−209−14) reporter, showing that the region between −209 and −14 is not involved in this effect (Fig. 4b). In addition, by performing ChIP assays in H1299 cells transfected with p53R280K, we were able to amplify a fragment between positions −2178 and −2071 on the ICMT promoter (ICMT up) upon immunoprecipitation with an anti-p53 antibody (Fig. 4c). In contrast, we could not amplify the fragment between positions −66 and +36 (ICMT down) on mutant p53-bound DNA (Fig. 4c). Therefore, our results show that p53R280K is recruited on the ICMT promoter on a different region than WT p53, indicating that each protein regulates promoter activity by different mechanisms.
Figure 4.
Effect of mutant p53 on the ICMT promoter. a, luciferase assays on promoter deletions. H1299 cells were co-transfected with the indicated reporters and a plasmid expressing p53R280K or empty vector (control). Luciferase activity was normalized to β-gal activity and expressed as arbitrary units (two-way ANOVA, ***, p < 0.001). b, luciferase assays on H1299 cells co-transfected with pICMTlucΔ(−209−14) and a plasmid expressing p53R280K or empty vector (control). Luciferase activity was normalized and expressed as fold change compared with control cells (one-tailed t test, p = 0.0256, *, p < 0.05). Lower panel, confirmation of p53R280K expression by Western blotting. c, ChIP assay on H1299 cells co-transfected with pICMTluc and pCDNA3-p53R280K or pCDNA3 as a control. IP was performed using anti-p53 antibody (DO1). The presence of the −2178 and − 2071 (ICMTup) or the −66 + 36 (ICMTdown) promoter fragments in immunoprecipitates was determined by qPCR (upper panel, n = 3, one-tailed t test, p = 0.0476) and semiquantitative PCR (lower panel).
ICMT overexpression enhances tumor phenotypes
Collectively, our results showed that the ICMT promoter is under negative regulation by the p53 pathway. Conversely, tumor-associated p53 mutants counteracted this repressive effect, suggesting that the acquisition of missense mutations on TP53 may further cooperate to increase ICMT expression. On this basis, we hypothesized that an uncontrolled increase in ICMT levels may cooperate with pro-oncogenic processes. Therefore, we decided to study the consequences of increasing ICMT levels on tumor-associated phenotypes. To this end, we generated the pLPC-ICMT-GFP retroviral construct, containing the ICMT coding sequence fused to the enhanced GFP N terminus. H1299 cells were transduced with retroviral particles containing pLPC-ICMT-GFP or pLPC-GFP as control. ICMT-GFP expression was confirmed by Western blotting and fluorescence microscopy (Fig. S2, a and b). We observed that ICMT-GFP localized in a discrete cytoplasmic area in close association with the nucleus, in agreement with the localization in the endoplasmic reticulum described for this enzyme (36). In contrast, GFP was uniformly distributed in the cytoplasm of control cells. We first analyzed the effect of ICMT on cell proliferation in vitro (Fig. S2c), but no significant differences in the proliferation rate between ICMT overexpressing cells and control cells were found.
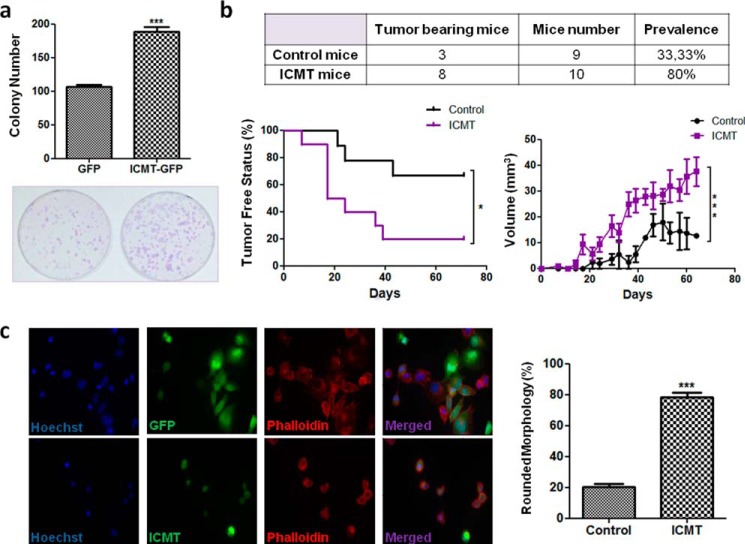
To understand whether high ICMT levels may promote clonogenic capacity in vitro, we performed colony formation assays. H1299 cells stably expressing pLPC-ICMT-GFP or pLPC-GFP were plated at low density and incubated to allow clonal growth. Our results showed that cells overexpressing ICMT developed a significantly larger number of colonies compared with control cells (Fig. 5a). These results suggest that high ICMT levels may confer an advantage to survive and proliferate under restrictive conditions, such as those imposed by low-density culture. Accordingly, treatment of cells with the ICMT inhibitor cysmethynil reduced the clonogenic potential of H1299 and MDA-MB-468 breast cancer cells in similar colony formation assays (Fig. S2d). To further explore this aspect, we studied the ability of ICMT-overexpressing cells to develop tumors in vivo (Fig. 5b). H1299 cells stably expressing ICMT-GFP, or GFP as control, were injected subcutaneously into nude mice, and tumor development was monitored periodically. We found that 80% of mice injected with cells expressing ICMT-GFP developed tumor masses (8/10). In contrast, tumors were found in only 33.3% of mice injected with control cells (3/9). Moreover, tumors arising from ICMT-GFP–expressing cells developed significantly earlier compared with controls. We also observed that palpable masses in control mice were smaller than those in mice injected with ICMT-GFP–expressing cells. In summary, our results showed that high ICMT levels enhanced the tumorigenic potential of H1299 cells in vivo.
Figure 5.
ICMT overexpression enhances tumor-associated phenotypes. a, colony formation assay. H1299 cells stably expressing ICMT-GFP or GFP as a control were plated at low density and incubated for 2 weeks, and the colonies were stained with Giemsa and the number of colonies determined (n = 4, one-tailed t test, p = 0.0003, ***, p < 0.005). b, in vivo tumor formation assay. H1299 cells stably expressing ICMT-GFP or GFP as a control were injected subcutaneously, and tumor development was monitored. Upper panel, number of mice that developed tumors in each group (ICMT or control). Lower left, Kaplan Meier graph showing the days when the masses were detectable by manual inspection after cell injection (log-rank (Mantel-Cox) test, p = 0.0240 and Gehan-Breslow-Wilcoxon test, p = 0.0177). Lower right, size evolution of the palpable masses detected in each group (comparison of fits, exponential growth equation, p < 0.0001). c, visualization of actin polymerization. H1299 cells transfected with pLPC-GFP (upper left panel) or pLPC-ICMT-GFP (lower left panel) were plated on Matrigel-coated coverslips and stained with phalloidin. Representative images are shown. Right panel, the number of rounded cells was quantified (n = 3, two-tailed t test, p = 0.0052).
Next, we wondered which are the mechanisms activated by ICMT overexpression. Some studies were focused on particular substrates such as RAS and RHO GTPases. Evidence from models of Ras-induced tumorigenesis has shown that ERK phosphorylation is reduced upon ICMT genetic inactivation (14). Therefore, to understand whether ICMT may have a similar effect in H1299 cells, we performed Western blotting studies with a p-ERK antibody. We found that phosphorylated ERK is readily detectable, suggesting a high level of this modification. This observation may be explained by the presence of a constitutively active NRAS mutant form in this cell line. Neither ICMT knockdown nor cysmethynil treatment affected ERK phosphorylation, which, on the contrary, was clearly reduced upon treatment with the MEK inhibitor U0126 (Fig. S2e). Therefore, our results strongly suggest that the pro-oncogenic effects of ICMT in H1299 cells do not involve ERK activation by RAS. To further explore the mechanism affected by ICMT, we studied changes on the cytoskeleton, in view of the relevance of RHO GTPases as regulators of actin polymerization. Cells were transfected with pLPC-ICMT-GFP or pLPC-GFP, plated on Matrigel-covered glass, and probed with fluorophore-conjugated phalloidin. Actin distribution was analyzed by fluorescence microscopy in GFP-positive cells. Upon transfection of ICMT-GFP, a large proportion of cells showed a rounded morphology with accumulation of actin near the plasma membrane, suggesting filament bundling. In contrast, the proportion of control cells showing this morphology was markedly reduced (Fig. 5c). These results showed that ICMT overexpression altered actin distribution in H1299 cells.
Interplay between ICMT and p53 status in breast and lung cancer
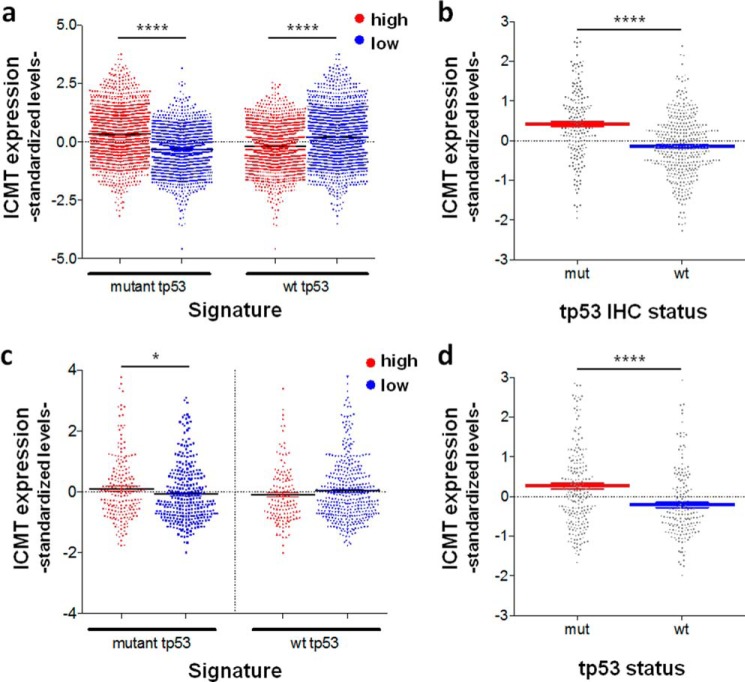
Our results from in vitro and in vivo experiments suggest that ICMT overexpression can cooperate to force mechanisms of aggressiveness, and consequently, its expression should be kept under strict control by proteins with tumor-suppressive functions, like WT p53. Conversely, the acquisition of mutations in TP53 may cooperate with tumor progression by promoting ICMT overexpression. To explore the clinical relevance of our findings, we analyzed breast cancer public databases. First, we wondered whether p53 status affects ICMT expression. We analyzed microarray data from 25 independent public databases searching for correlations between ICMT mRNA levels and p53 function (Breast Cancer compendium: 3661 cases). Because TP53 mutation status was not available in this compendium, we analyzed the expression of the p53 gene signature, to discriminate between cases with WT p53 and those with p53 alterations. The WT p53 signature was defined based on the expression profile of human tumors with known p53 status and was previously shown to be strongly correlated with the presence of WT p53 alleles (37). Conversely, the mutant p53 signature was correlated with the presence of a mutant TP53 gene, including cases with different types of mutations (deletions, nonsense, and missense). We found that cases with a high mutant p53 signature score showed relatively higher ICMT mRNA levels. Accordingly, cases with a high WT p53 signature score showed significantly lower levels of ICMT mRNA (Fig. 6a). These results strongly suggest that the presence of WT p53 in tumors correlates with low ICMT expression, in line with our results from experimental models. Taking into account that most cases with TP53 mutation are likely to express p53 point mutants, our results also suggest that mutant p53 favors ICMT overexpression in tumors. High p53 protein levels in tumors are strongly correlated with the presence of p53 mutant forms (38). Therefore, we analyzed ICMT mRNA levels in cases where data of p53 expression, assessed by immunohistochemistry (IHC), was available (596 cases). Supporting our observations, ICMT mRNA levels significantly increased in cases with high p53 protein levels (Fig. 6b).
Figure 6.
Expression levels of ICMT in breast and lung cancer. Standardized expression levels of ICMT in samples from the breast cancer compendium (n = 3661) stratified according to high and low mutant p53 and high and low WT p53 expression signature score (see text) (a) and p53 IHC status (b). Standardized expression levels of ICMT in lung adenocarcinoma samples from the TCGA-LUAD dataset (n = 516) stratified according to high and low mutp53 and high and low WT p53 expression signature score (c) and p53 status as determined from whole-exome sequencing (****, p value < 0.0001, and *, p value < 0.05 in a two-tailed unpaired t test) (d).
Next, we extended our analysis to determine whether similar correlations may be observed in lung cancer. To this end, we classified cases from the TCGA lung adenocarcinoma dataset (LUAD: 516 cases) according to the expression of the p53 signature, as explained for the BC compendium. We found significantly higher ICMT mRNA levels in cases with a high mutant p53 signature score as compared with cases with a low mutant p53 signature score (Fig. 6c). Accordingly, the mean standardized level of ICMT expression was reduced by more than 2-fold in cases with a high WT p53 signature score (Fig. S3). To further explore the correlation between ICMT expression and the p53 pathway in lung cancer, we analyzed mRNA levels in cases with TP53 mutations, assessed by whole-exome sequencing, because p53 IHC data are not available in this dataset. In support of our hypothesis, we found that ICMT mRNA levels were significantly higher in cases bearing TP53 mutations, compared with WT p53 cases (Fig. 6d).
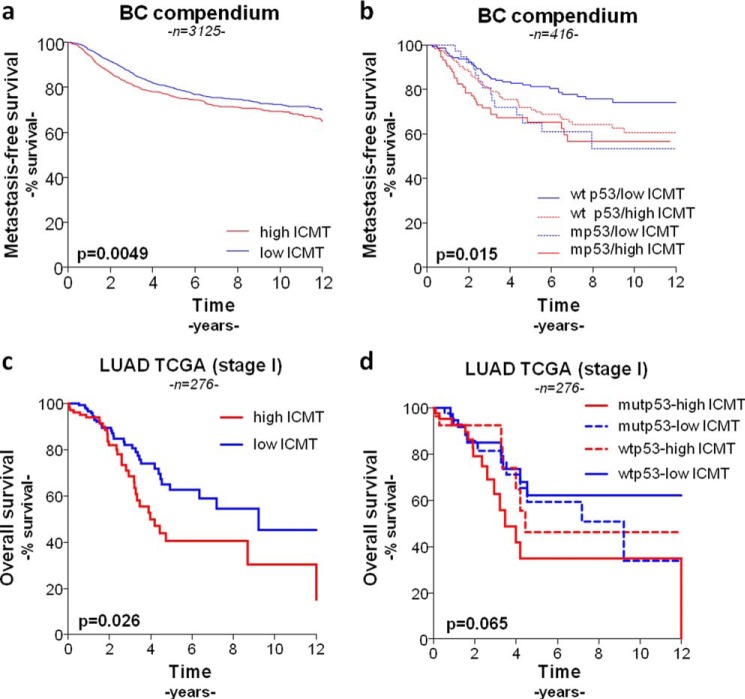
Finally, we wondered whether ICMT expression may correlate with clinical outcome. We found that cases with high ICMT mRNA levels in the BC compendium showed a significant decrease in metastasis-free survival (Fig. 7a), suggesting that ICMT overexpression cooperates with metastasis development in breast cancer. When we stratified cases considering also p53 expression levels determined by IHC, we found that cases with low p53 expression (i.e. correlated with WT p53 status) and low ICMT mRNA levels showed a significantly increased metastasis-free survival compared with the other groups (Fig. 7b). These results suggest that the metastasis-promoting effect of ICMT is stronger in tumors retaining WT p53. Likewise, we found a significant correlation between high ICMT expression levels and a decreased overall survival in the lung cancer dataset (Fig. 7c). A similar tendency as that observed in the BC compendium was found when we stratified cases considering p53 expression levels (Fig. 7d). Collectively, our results support the idea that ICMT overexpression is associated with p53 pathway alteration and promotes tumor aggressiveness in breast and lung cancer.
Figure 7.
Survival analysis of breast cancer and stage I lung adenocarcinoma samples stratified according to ICMT and p53 status. Metastasis-free survival of breast cancer samples from the breast cancer compendium (n = 3661) stratified according to high and low ICMT expression level (a); high and low ICMT expression level and p53 IHC status (WT p53, low p53 expression; mp53, high p53 expression) (b). Overall survival of stage I TCGA-LUAD samples (n = 276) stratified according to high and low ICMT expression level (c); high and low ICMT expression level and p53 status (d) (log-rank Mantel-Cox) test).
Discussion
Protein prenylation is emerging as a critical post-translational modification that affects different aspects of cell physiology. Recently, exciting evidence proposed that altered prenylation or post-prenylation processing may cooperate with pathologies such as chronic inflammation, cancer, progeria, and neurological disorders (7). However, the underlying mechanisms are not completely understood. The existence of three modification steps makes the study of protein prenylation and its biological consequences a challenging task, but it also provides additional opportunities to manipulate the process (7). Inhibition of farnesyl transferase was proposed as a therapeutic strategy in cancer; however, the molecules tested in clinical trials showed a limited response. A possible explanation to this disappointing performance may be alternative prenylation by geranylgeranyl transferase I. Targeting post-prenylation processing offers the advantage that both RCE1 and ICMT can act on either farnesylated or geranylgeranylated substrates. Studies on RCE1 showed relatively modest results and possible adverse effects, including cardiomyopathy and retinopathy (7). In contrast, encouraging results for ICMT inactivation or inhibition have been observed in some experimental models. Therefore, a profound knowledge of the alteration of ICMT function in tumors is necessary to exploit its potential as a therapeutic target.
Studies in mice suggested that ICMT is differentially expressed, showing lower mRNA levels in skeletal muscle compared with brain and liver (39). Besides, a reduction in ICMT mRNA levels by binding of miR100 was observed in hepatocarcinoma cell lines (40). Nevertheless, to our knowledge, there are no further studies on ICMT expression and, in particular, on transcriptional regulation. In this work, we showed that the ICMT promoter is under regulation by p53 forms, including tumor-associated p53 point mutants, suggesting a complex interplay that may affect cell physiology.
Our results show for the first time that ICMT is a target for transcriptional repression by WT p53. The effects exerted by WT p53 on gene expression are complex and may be influenced by cell context and the nature of the activating signals. Nevertheless, several independent studies have firmly established that WT p53 not only up-regulates but also down-regulates specific target genes (41). Different models were proposed to explain repression by WT p53, including direct and indirect mechanisms. We identified the region between −209 and −14 as responsible for the observed effect. Moreover, we found that WT p53 is recruited to the ICMT promoter in a region encompassing this fragment, strongly suggesting that the mechanism of repression involves the action of WT p53 as part of a regulatory complex on chromatin. In silico analysis of the identified promoter region failed to identify a p53 response element, even considering a modified consensus proposed to account for the deviations found in promoters of repressed target genes (42). Thus, we speculate that recruitment may involve the interaction with other proteins present in the promoter, although we cannot exclude binding to a highly divergent site.
In contrast, we showed that several p53 point mutants enhance ICMT expression, suggesting that overexpression of this enzyme may be related to mutant p53 oncogenic function. Our evidence also shows that this effect is associated with a gain-of-function mechanism, because the enhancement of reporter activity and ICMT protein levels in H1299 cells is completely independent from WT p53. Moreover, our results showing that the effect of p53R280K was independent of the −209 and −14 region and that the mutant protein was recruited on a different region on the promoter indicate that WT and mutant p53 act through different mechanisms. In addition, co-expression with p53R280K eliminated the repressive effect of WT p53, showing a dominant-negative effect. Thus, our results imply that the acquisition of missense mutations on TP53 may cooperate to increase ICMT expression by complementary mechanisms. On the one hand, the repressive function of WT p53 may be lost upon mutation of one TP53 allele and inhibition of the remaining WT protein by mutant p53. On the other hand, point mutants may activate WT p53-independent mechanisms.
Our findings suggest that ICMT expression should be strictly regulated in physiological conditions while the alterations acquired during tumorigenesis, leading to functional inactivation of WT p53, would induce ICMT expression. In this context, such an increase in ICMT levels would be expected to cooperate with tumor progression. Supporting this idea, we show for the first time that ICMT overexpression enhances clonogenicity in vitro as well as tumorigenic potential in vivo. Some studies have described the effects of ICMT inhibition or silencing; however, the effects of its overexpression have been poorly studied. We provide original evidence showing that ICMT overexpression enhances tumor-associated phenotypes in non-small cell lung carcinoma H1299 cells, suggesting that its deregulation may be relevant in this cancer type. Our results showed that the actin cytoskeleton is affected by ICMT overexpression. Considering the key role played by RHO GTPases as regulators of actin polymerization, it is likely that their activity may be affected by changes on ICMT levels. Individual overexpression of RHOA, RAC1, or CDC42 in the same conditions did not recapitulate the effect caused by ICMT (data not shown). Therefore, our results failed to identify a particular RHO GTPase as responsible for the observed effect. Alternatively, the alteration of actin cytoskeleton may be explained by the simultaneous action on several RHO GTPases.
Our analysis of public databases provides support to the idea that WT and mutant p53 exert opposite effects on ICMT expression in breast cancer, showing that tumors classified as WT p53 are correlated with lower ICMT mRNA levels. Conversely, tumors classified as mutant p53 showed significantly higher ICMT mRNA levels. In addition, this hypothesis is further supported by our analysis based on p53 expression. Although p53 levels cannot be considered as direct proof of p53 mutation, there is a strong association between p53 mutation and p53 overexpression in tumors (38). Our results showed a marked correlation between tumors with high ICMT mRNA levels and p53 overexpression. Considering that missense mutations are the most frequent genetic alterations on TP53, this evidence is in line with our in vitro results showing that p53 point mutants enhance ICMT expression. A similar correlation between ICMT mRNA levels and p53 status was observed in the lung cancer dataset, suggesting that the effect of the p53 pathway on ICMT expression is not restricted to a specific tumor type.
In agreement with our results from experimental models, ICMT overexpression showed a significant correlation with poor prognosis in the breast and lung cancer datasets analyzed. A more detailed analysis of the BC compendium indicated that cases with low p53 and low ICMT expression displayed a significantly increased metastasis-free survival compared with all other groups. According to the criteria used in this classification, the group with low p53 expression is expected to include most of the WT p53 cases and all the p53 null cases. Consequently, our results suggest that ICMT overexpression in tumors that do not express mutant p53 forms (i.e. WT or null) would have a stronger effect on metastasis development, compared with cases expressing p53 mutants. On this basis, it is possible to hypothesize that preventing ICMT overexpression, or inhibiting its function, in the low p53 expression group would increase survival. These results also suggest that therapies based on ICMT inhibition may be particularly efficient in patients with WT p53, which represent more than 70% of breast cancer cases.
Previous evidence has shown that enhanced protein geranylgeranylation cooperates with tumor aggressiveness in breast cancer and that p53 point mutants promote this effect by altering the mevalonate pathway (43, 44). ICMT overexpression may recapitulate some of the effects of mevalonate pathway alteration by affecting the function of geranylgeranylated proteins. In this context, mutant p53 tumors may be less dependent on ICMT overexpression because they have already developed a mechanism to alter the function of proteins modified by geranylgeranylation. This partially overlapping effect may explain why ICMT overexpression exerted a more marked effect on the low p53 expression group (enriched in WT p53 cases) of the BC compendium (Fig. 7b). Furthermore, expression of p53 point mutants are also predicted to activate other mechanisms of tumor aggressiveness that will be absent in WT or null p53 cases (30). Therefore, mutant p53-activated mechanisms would provide additional driving forces to foster tumor progression in cases with low ICMT levels. This could help us to understand why ICMT expression did not show a significant difference in survival among cases in the high p53 expression group (enriched in mutant p53 cases, Fig. 7b).
In summary, our results unveil a connection between the p53 pathway and the prenylated protein network. We propose that ICMT expression is repressed by WT p53. Alteration of this regulation during tumor progression would impact the ICMT levels, which in turn would modify the action of prenylated proteins. Our findings reveal a mechanism through which tumor cells could manipulate the regulation of prenylated proteins to foster mechanisms of aggressiveness. Moreover, our findings also contribute to understanding the clinical relevance of ICMT overexpression in breast and lung cancer and suggest that therapeutic strategies based on ICMT inhibition may be particularly useful in WT p53 cases.
Experimental procedures
Cell culture
MDA-MB-231, MDA-MB-468, MCF-7, HT29, HEK293-GP, HCT116 p53+/+, and HCT116 p53−/− were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 units/ml penicillin and streptomycin (Invitrogen). H1299 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 100 units/ml penicillin and streptomycin (Invitrogen). Cell lines were purchased from ATCC, and authenticity was documented by standard STR analysis. Cells were cultured in a humidified incubator at 37 °C with 5% CO2 and tested periodically for mycoplasma by 4′,6-diamidino-2-phenylindole staining and PCR.
Cell transfection and retroviral transduction
DNA and siRNA transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The following siRNAs sequences were used: sip53 targeting human TP53 (GACUCCAGUGGUAAUCUAC), siLacZ targeting the lacZ gene from Escherichia coli (GUGACCAGCGAAUACCUGU) (25), and siICMT (SMART pool M-005209-01-0005, Dharmacon). Stable genetic manipulation was performed by transduction with retrovirus-based plasmids as described previously (25). The following shRNAs were used for stable knockdown (25): shp53 (GACUCCAGUGGUAAUCUAC), shp53 3′UTR (GGUGAACCUUAGUACCUAA), and shLacZ (GUGACCAGCGAAUACCUGU).
Gene expression analysis
Total RNA was extracted using TRIzol (Invitrogen) and subjected to DNase I (Promega) treatment. RNA was retro-transcribed using Moloney murine leukemia virus retrotranscriptase (Promega). Real-time PCR was performed using SYBR Green PCR master mix (Biodynamic) according to the following conditions: 2 min at 95 °C for one cycle; and 30 s at 95 °C, 20 s at 60 °C, and 30 s at 72 °C for 40 cycles. Results were analyzed using the comparative ΔΔCt method. Values were normalized to GAPDH expression. The following real time PCR primers were used: ICMTqPfw; ICMTqPrv; GAPDHfw; GAPDHrv; ICMTPrtfw-ChIP; ICMTPrtrv-ChIP; ICMTPrtfw-ChIPup; and ICMTPrtrv-ChIPup (Table S1).
Plasmids
Plasmid DNA was prepared using Wizard DNA purification kit (Promega). For WT and mutant p53 transient expression pCDNA3-p53 (45), pCDNA3-p53R280K (25), pCDNA3-p53R273H, pCDNA3-p53R175H, and pCDNA3-p53R248W, (25) plasmids were used. pCDNA3-p53R249S was generated by site-directed mutagenesis on pCDNA3-p53. pLPC-ICMT-GFP was constructed using pLPC-GFP vector (46). ICMT coding sequence was amplified by PCR on cDNA from H1299 cells using ICMTuppLFw and ICMTpLRv. To generate the pICMTluc reporter, a fragment spanning from −2234 to +37 on the ICMT promoter was amplified by PCR on genomic DNA from MDA-MB-231 cells using ICMP2fw and ICMP1rv as primers and cloned into the pGL3-basic vector. This construct was used as a PCR template to generate a series of 5′-terminally truncated ICMT promoter fragments: ICMTluc1000 (primers: ICMPMfw and ICMP1rv); ICMTluc500 (primers: ICMTQSacIfw and ICMP1rv); ICMTluc200 (primers: ICMTP200SacIfw and ICMP1rv); and ICMTluc50 (primers: ICMTP50SacIfw and ICMP1rv). Each fragment was ligated into pGL3 basic vector. pICMTlucΔ(−209 and −14) was generated by overlap extension PCR using the following primer pairs: ICMP2fw/ICMTPDrv (upstream fragment) and ICMTPDfw/ICMP1rv (downstream fragment). Fragments were annealed and used as template to amplify a 2077-bp fragment with ICMP2fw and ICMP1rv primers. The fragment was cloned into pGL3 vector. To generate the pGL3P-ICMTluc200-50 reporter, a fragment including the region between positions −209 and −14 on the ICMT promoter was amplified by PCR using pICMTluc as a template (primers: ICMTP200KpnIfw and ICMTP50upSacIrv) and cloned into pGL3-promoter vector (primer sequences are shown in Table S1).
Luciferase reporter assay
H1299 and HCT116 cells were co-transfected with the indicated plasmids and pCMV-β-gal (Promega) as a control of transfection efficiency, using Lipofectamine 2000 (Invitrogen). Transfected cells were harvested in 1× Passive Lysis Buffer (Promega). Luciferase activity was measured using Luciferase Assay Reagent (Promega) in a Multi-Mode Microplate Reader (SynergyTM 2, BioTek). All experiments were repeated at least three times. The values were normalized relative to β-gal activity. When indicated, the HCT116 cells were treated with 0.5 μm doxorubicin (Sigma) for 16 h.
ChIP assay
ChIP assays were performed as described previously (25). Cells were transfected with pCDNA3-WTp53, pCDNA3-p53R280K, or pCDNA3 as a control and pICMTluc. Chromatin was sonicated to 500–800-bp average fragment size and pre-cleared for 1 h at 4 °C with protein A-Sepharose (GE Healthcare). Chromatin was immunoprecipitated with p53 DO1 (Santa Cruz Biotechnology). Co-immunoprecipitated DNA was analyzed by real time PCR. Promoter occupancy was calculated as percent of input chromatin using the ΔΔCt method.
Colony formation assay
H1299 cells stably expressing ICMT-GFP or GFP as a control were generated by retroviral transduction. Cells were plated at low density in 60-mm plates and cultured for 15 days to allow colony formation. Colonies were stained with Giemsa solution (Biopur Diagnostics) and counted. To evaluate the effect of cysmethynil (Cayman Chemical), H1299 or MDA-MB-468 cells were plated at low density in 60-mm plates and treated with 30 μm cysmethynil. For treatments, a stock solution of 13 mm cysmethynil in DMSO was diluted in culture medium, and the corresponding dilution of DMSO was used as control.
Western blotting and antibodies
Western blotting was performed as described previously (25). As primary antibodies, anti-GFP Ab290 (Abcam), anti-p53 DO-1 (Santa Cruz Biotechnology), anti-ICMT (Proteintech, 51001-2-AP), anti-actin (Sigma, A2066), anti-GAPDH G-9 (Santa Cruz Biotechnology), ERK C-9 (Santa Cruz Biotechnology), and p-ERK (Cell Signaling, 91015) were used. HRP-conjugated anti-rabbit (The Jackson Laboratory, 111-035-003) and HRP-conjugated anti-mouse (The Jackson Laboratory, 115-035003) antibodies were used as secondary antibodies. Chemiluminescence was detected using Amersham Biosciences ECL Prime Western blotting detection reagent (GE Healthcare). For WT p53 stabilization, MCF7 or HCT116 p53+/+ cells were treated with 10 μm Nutlin-3 (Sigma) for 48 h.
In vivo tumor formation
Procedures involving animals conformed to institutional guidelines that comply with international laws and policies (Council for International Organization of Animal Sciences (CIOMS) and International Council for Laboratory Animal Science (ICLAS)). All experimental protocols were approved by the Animal Ethics Committee of the National University of Rosario (CICUAL-FBioyF). Six- to 8-week-old female nude mice were obtained from the Animal facility of the University of La Plata, Argentina. Animals were fed commercial chow and water ad libitum and maintained in a 12-h light/dark cycle. Mice were randomly divided into two groups; group I, control (n = 9), was injected with H1299 cells stably expressing GFP, and group II, ICMT (n = 10) was injected with H1299 cells stably expressing ICMT. Cells were diluted 1:1 in Matrigel (Corning) and injected subcutaneously (1 × 106/mouse). Palpable masses were measured twice a week with a caliper to determine major (D) and minor (d) diameters, and the volume calculated using the ellipsoidal formula V = π/6 × D × d2.
F-actin staining
H1299 cells were transfected with pLPC-GFP or pLPC-ICMT-GFP and 16 h later were plated in coverslips pre-coated with 1:100 Matrigel (Corning). After 24 h, cells were fixed and stained with 1:200 fluorescent phalloidin (Life Technologies, Inc.). On average, 25 photos were taken per condition, and the experiment was performed three times. Rounded cells with strong F-actin signal in proximity of the plasma membrane were counted over total cells.
Cell proliferation
H1299 cells were seeded at low density (5 × 104) on 35-mm culture plates and incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cell number was determined at different time points using a Neubauer chamber.
Collection and processing of gene expression data
Breast cancer
We started from a collection of 4640 samples from 27 major data sets comprising microarray data of breast cancer samples annotated with histological tumor grade and clinical outcome. All data were measured on Affymetrix arrays and have been downloaded from NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and EMBL-EBI ArrayExpress (http://www.ebi.ac.uk/arrayexpress/)3 (49). Prior to analysis, we re-organized all datasets eliminating duplicate samples and re-naming each original set after the medical center where the patients were recruited. Briefly, the datasets have been modified as described in Ref. 47.
Lung adenocarcinoma
Gene expression RSEM (RNA-Seq by Expectation-Maximization) level 3 normalized data, TP53 mutations, and clinical information for n = 516 samples of the TCGA LUAD dataset were downloaded from the GDAC Firehose.
MDA-MB-231 cancer cell lines
Raw gene expression data (.CEL files) for control and mutant p53-depleted MDA-MB-231 cells were downloaded from NCBI Gene Expression Omnibus GSE14491. Probe level signals were converted to expression values using robust multi-array average procedure RMA (48) of Bioconductor affy package.
Average signature expression and signature scores
Average signature expression was calculated as the standardized average expression of all signature genes in sample subgroups (e.g. p53 signatures; p53 IHC status, ICMT 201609_x_at probe set). Signature scores were obtained summarizing the standardized expression levels of signature genes into a combined score with zero mean (32). The values shown in graphs are thus dimensionless.
Kaplan-Meier survival analysis
To identify two groups of tumors with either high or low ICMT signature, we used the classifier described in Ref. 32 that is a classification rule based on the ICMT signature score (201609_x_at probe set). Tumors were classified as ICMT signature “Low” if the combined score was negative and as ICMT signature “High” if the combined score was positive. This classification was applied to expression values of the metadataset. To evaluate the prognostic value of the ICMT signature, we estimated, using the Kaplan-Meier method, the probabilities that patients would remain free of metastasis To confirm these findings, the Kaplan-Meier curves were compared using the log-rank (Mantel-Cox) test. p values were calculated according to the standard normal asymptotic distribution. Survival analysis was performed in GraphPad Prism.
Author contributions
C. B. E. and J. G. conceptualization; C. B. E., S. B., and J. G. data curation; C. B. E., C. D. B., and S. B. software; C. B. E., C. D. B., S. B., M. M.-M., and J. G. formal analysis; C. B. E., S. B., and J. G. validation; C. B. E., C. D. B., C. R., M. V. B., S. B., M. M.-M., and J. G. investigation; C. B. E., C. D. B., M. V. B., S. B., G. D. S., and J. G. visualization; C. B. E., C. D. B., S. B., M. M.-M., and J. G. methodology; C. B. E., C. D. B., S. B., G. D. S., and J. G. writing-review and editing; G. D. S., M. M.-M., and J. G. resources; J. G. supervision; J. G. funding acquisition; J. G. writing-original draft; J. G. project administration.
Supplementary Material
Acknowledgments
We are grateful to Cecilia Farre and Gustavo Chapo from the Medical School Animal Facility of the National University of Rosario for help with mouse housing and monitoring. We thank Cecilia Larocca and Evangelina Almada for helpful discussion and assistance with cytoskeleton analysis. We thank the staff from the English Department of the Facultad de Ciencias Bioquímicas y Farmacéuticas (UNR) for the language correction of the manuscript.
This work was supported by Instituto Nacional del Cáncer (INC) Asistencia Financiera III, 2015 grants (to J. G.) and Agencia Nacional de Promoción Científica y Tecnológica Grant ANPCyT. PICT 1221. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Table S1.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ICMT
- isoprenyl cysteine carboxyl methyltransferase
- IHC
- immunohistochemistry
- qRT
- quantitative RT
- qPCR
- quantitative PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- IP
- immunoprecipitation
- ANOVA
- analysis of variance
- ERK
- extracellular signal–regulated kinase
- LUAD
- lung adenocarcinoma.
References
- 1. Liang P. H., Ko T. P., and Wang A. H. (2002) Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269, 3339–3354 10.1046/j.1432-1033.2002.03014.x [DOI] [PubMed] [Google Scholar]
- 2. Smeland T. E., Seabra M. C., Goldstein J. L., and Brown M. S. (1994) Geranylgeranylated Rab proteins terminating in Cys-Ala-Cys, but not Cys-Cys, are carboxyl-methylated by bovine brain membranes in vitro. Proc. Natl. Acad. Sci. U.S.A. 91, 10712–10716 10.1073/pnas.91.22.10712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manolaridis I., Kulkarni K., Dodd R. B., Ogasawara S., Zhang Z., Bineva G., O'Reilly N. O., Hanrahan S. J., Thompson A. J., Cronin N., Iwata S., and Barford D. (2013) Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature 504, 301–305 10.1038/nature12754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winter-Vann A. M., and Casey P. J. (2005) Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 5, 405–412 10.1038/nrc1612 [DOI] [PubMed] [Google Scholar]
- 5. Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F. L., and Eisenhaber F. (2007) Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 3, e66 10.1371/journal.pcbi.0030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen U. T., Guo Z., Delon C., Wu Y., Deraeve C., Fränzel B., Bon R. S., Blankenfeldt W., Goody R. S., Waldmann H., Wolters D., and Alexandrov K. (2009) Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat. Chem. Biol. 5, 227–235 10.1038/nchembio.149 [DOI] [PubMed] [Google Scholar]
- 7. Wang M., and Casey P. J. (2016) Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17, 110–122 10.1038/nrm.2015.11 [DOI] [PubMed] [Google Scholar]
- 8. Hancock J. F., Magee A. I., Childs J. E., and Marshall C. J. (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–1177 10.1016/0092-8674(89)90054-8 [DOI] [PubMed] [Google Scholar]
- 9. Casey P. J., Solski P. A., Der C. J., and Buss J. E. (1989) p21ras is modified by a farnesyl isoprenoid. Proc. Natl. Acad. Sci. U.S.A. 86, 8323–8327 10.1073/pnas.86.21.8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown M. S., and Goldstein J. L. (1980) Multivalent feedback regulation of HMG-CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 21, 505–517 [PubMed] [Google Scholar]
- 11. Quesney-Huneeus V., Wiley M. H., and Siperstein M. D. (1979) Essential role for mevalonate synthesis in DNA replication. Proc. Natl. Acad. Sci. U.S.A. 76, 5056–5060 10.1073/pnas.76.10.5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Habenicht A. J., Glomset J. A., and Ross R. (1980) Relation of cholesterol and mevalonic acid to the cell cycle in smooth muscle and Swiss 3T3 cells stimulated to divide by platelet-derived growth factor. J. Biol. Chem. 255, 5134–5140 [PubMed] [Google Scholar]
- 13. Bergo M. O., Gavino B. J., Hong C., Beigneux A. P., McMahon M., Casey P. J., and Young S. G. (2004) Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J. Clin. Invest. 113, 539–550 10.1172/JCI200418829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wahlstrom A. M., Cutts B. A., Liu M., Lindskog A., Karlsson C., Sjogren A. K., Andersson K. M., Young S. G., and Bergo M. O. (2008) Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood 112, 1357–1365 10.1182/blood-2007-06-094060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau H. Y., Tang J., Casey P. J., and Wang M. (2017) Isoprenylcysteine carboxylmethyltransferase is critical for malignant transformation and tumor maintenance by all RAS isoforms. Oncogene 36, 3934–3942 10.1038/onc.2016.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Court H., Amoyel M., Hackman M., Lee K. E., Xu R., Miller G., Bar-Sagi D., Bach E. A., Bergö M. O., and Philips M. R. (2013) Isoprenylcysteine carboxylmethyltransferase deficiency exacerbates KRAS-driven pancreatic neoplasia via Notch suppression. J. Clin. Invest. 123, 4681–4694 10.1172/JCI65764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cushman I., and Casey P. J. (2011) RHO methylation matters: a role for isoprenylcysteine carboxylmethyltransferase in cell migration and adhesion. Cell Adh. Migr. 5, 11–15 10.4161/cam.5.1.13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jansen S., Gosens R., Wieland T., and Schmidt M. (2018) Paving the Rho in cancer metastasis: Rho GTPases and beyond. Pharmacol. Ther. 183, 1–21 10.1016/j.pharmthera.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 19. Cushman I., and Casey P. J. (2009) Role of isoprenylcysteine carboxylmethyltransferase-catalyzed methylation in Rho function and migration. J. Biol. Chem. 284, 27964–27973 10.1074/jbc.M109.025296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Do M. T., Chai T. F., Casey P. J., and Wang M. (2017) Isoprenylcysteine carboxylmethyltransferase function is essential for RAB4A-mediated integrin β3 recycling, cell migration and cancer metastasis. Oncogene 36, 5757–5767 10.1038/onc.2017.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zandvakili I., Lin Y., Morris J. C., and Zheng Y. (2017) Rho GTPases: anti-or pro-neoplastic targets? Oncogene 36, 3213–3222 10.1038/onc.2016.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Backlund P. S. (1997) Post-translational processing of RhoA. Carboxyl methylation of the carboxyl-terminal prenylcysteine increases the half-life of RhoA. J. Biol. Chem. 272, 33175–33180 10.1074/jbc.272.52.33175 [DOI] [PubMed] [Google Scholar]
- 23. Roberts P. J., Mitin N., Keller P. J., Chenette E. J., Madigan J. P., Currin R. O., Cox A. D., Wilson O., Kirschmeier P., and Der C. J. (2008) Rho family GTPase modification and dependence on CAAX motif-signaled post-translational modification. J. Biol. Chem. 283, 25150–25163 10.1074/jbc.M800882200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gentry L. R., Nishimura A., Cox A. D., Martin T. D., Tsygankov D., Nishida M., Elston T. C., and Der C. J. (2015) Divergent roles of CAAX motif-signaled post-translational modifications in the regulation and subcellular localization of Ral GTPases. J. Biol. Chem. 290, 22851–22861 10.1074/jbc.M115.656710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Girardini J. E., Napoli M., Piazza S., Rustighi A., Marotta C., Radaelli E., Capaci V., Jordan L., Quinlan P., Thompson A., Mano M., Rosato A., Crook T., Scanziani E., Means A. R., et al. (2011) A Pin1/Mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20, 79–91 10.1016/j.ccr.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 26. Kastenhuber E. R., and Lowe S. W. (2017) Putting p53 in context. Cell 170, 1062–1078 10.1016/j.cell.2017.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kandoth C., McLellan M. D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J. F., Wyczalkowski M. A., Leiserson M. D. M., Miller C. A., Welch J. S., Walter M. J., Wendl M. C., et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soussi T., and Wiman K. G. (2007) Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell 12, 303–312 10.1016/j.ccr.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 29. Muller P. A., and Vousden K. H. (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freed-Pastor W. A., and Prives C. (2012) Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286 10.1101/gad.190678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Girardini J. E., Walerych D., and Del Sal G. (2014) Cooperation of p53 mutations with other oncogenic alterations in cancer. Subcell. Biochem. 85, 41–70 10.1007/978-94-017-9211-0_3 [DOI] [PubMed] [Google Scholar]
- 32. Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M. B., Guzzardo V., Parenti A. R., Rosato A., Bicciato S., Balmain A., and Piccolo S. (2009) A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 10.1016/j.cell.2009.01.039 [DOI] [PubMed] [Google Scholar]
- 33. Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., and Olivier M. (2016) TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum. Mutat. 37, 865–876 10.1002/humu.23035 [DOI] [PubMed] [Google Scholar]
- 34. Muller P. A., and Vousden K. H. (2013) p53 mutations in cancer. Nat. Cell Biol. 15, 2–8 10.1038/ncb2641 [DOI] [PubMed] [Google Scholar]
- 35. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., and Liu E. A. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- 36. Dai Q., Choy E., Chiu V., Romano J., Slivka S. R., Steitz S. A., Michaelis S., and Philips M. R. (1998) Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J. Biol. Chem. 273, 15030–15034 10.1074/jbc.273.24.15030 [DOI] [PubMed] [Google Scholar]
- 37. Miller L. D., Smeds J., George J., Vega V. B., Vergara L., Ploner A., Pawitan Y., Hall P., Klaar S., Liu E. T., and Bergh J. (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U.S.A. 102, 13550–13555 10.1073/pnas.0506230102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soussi T., and Béroud C. (2001) Assessing TP53 status in human tumours to evaluate clinical outcome. Nat. Rev. Cancer 1, 233–240 10.1038/35106009 [DOI] [PubMed] [Google Scholar]
- 39. Bergo M. O., Leung G. K., Ambroziak P., Otto J. C., Casey P. J., Gomes A. Q., Seabra M. C., and Young S. G. (2001) Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J. Biol. Chem. 276, 5841–5845 10.1074/jbc.C000831200 [DOI] [PubMed] [Google Scholar]
- 40. Zhou H. C., Fang J. H., Luo X., Zhang L., Yang J., Zhang C., and Zhuang S. M. (2014) Downregulation of microRNA-100 enhances the ICMT-Rac1 signaling and promotes metastasis of hepatocellular carcinoma cells. Oncotarget 5, 12177–12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan K. D., Galbraith M. D., Andrysik Z., and Espinosa J. M. (2018) Mechanisms of transcriptional regulation by p53. Cell Death Differ. 25, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang B., Xiao Z., Ko H. L., and Ren E. (2010) The p53 response element and transcriptional repression. Cell Cycle 9, 870–879 10.4161/cc.9.5.10825 [DOI] [PubMed] [Google Scholar]
- 43. Freed-Pastor W. A., Mizuno H., Zhao X., Langerød A., Moon S. H., Rodriguez-Barrueco R., Barsotti A., Chicas A., Li W., Polotskaia A., Bissell M. J., Osborne T. F., Tian B., Lowe S. W., Silva J. M., et al. (2012) Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 10.1016/j.cell.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sorrentino G., Ruggeri N., Specchia V., Cordenonsi M., Mano M., Dupont S., Manfrin A., Ingallina E., Sommaggio R., Piazza S., Rosato A., Piccolo S., and Del Sal G. (2014) Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 16, 357–366 10.1038/ncb2936 [DOI] [PubMed] [Google Scholar]
- 45. Mantovani F., Tocco F., Girardini J., Smith P., Gasco M., Lu X., Crook T., and Del Sal G. (2007) The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 14, 912–920 10.1038/nsmb1306 [DOI] [PubMed] [Google Scholar]
- 46. Guida E., Bisso A., Fenollar-Ferrer C., Napoli M., Anselmi C., Girardini J. E., Carloni P., and Del Sal G. (2008) Peptide aptamers targeting mutant p53 induce apoptosis in tumor cells. Cancer Res. 68, 6550–6558 10.1158/0008-5472.CAN-08-0137 [DOI] [PubMed] [Google Scholar]
- 47. Enzo E., Santinon G., Pocaterra A., Aragona M., Bresolin S., Forcato M., Grifoni D., Pession A., Zanconato F., Guzzo G., Bicciato S., and Dupont S. (2015) Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 34, 1349–1370 10.15252/embj.201490379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., and Speed T. P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T., Megy K., Pilicheva E., Rustici G., Tikhonov A., Parkinson H., et al. (2015) ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 43, D1113–D1116 10.1093/nar/gku1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.