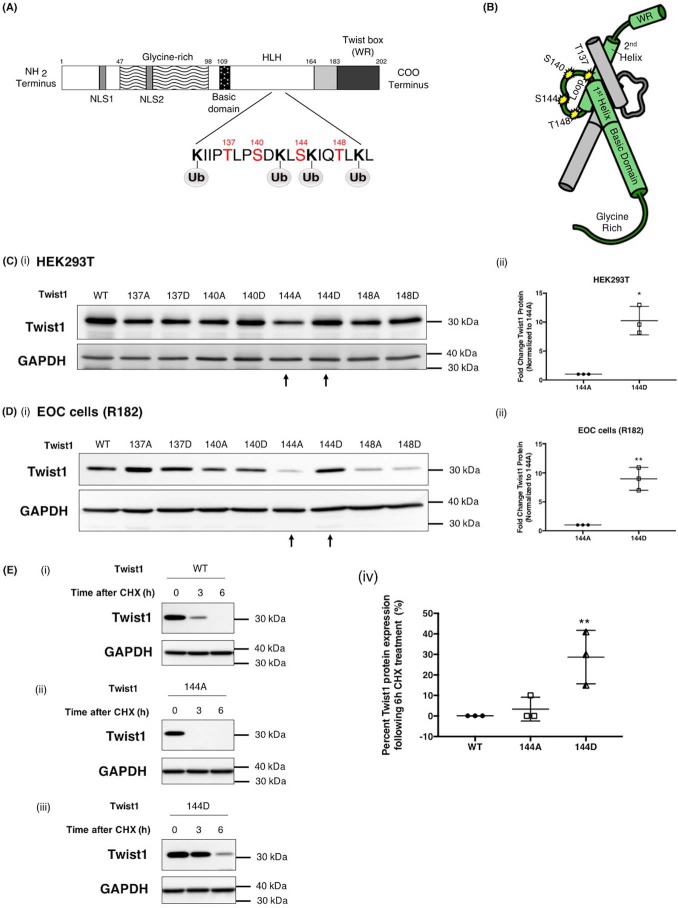
Figure 4.
Phosphorylation on Ser-144 of Twist1 promotes its stability. A, schematic representation of predicted PKCα phosphorylation sites on Twist1 adjacent to predicted Twist1 ubiquitination sites. B, location of phosphorylation sites included in this study (Thr-137, Ser-140, Ser-144, and Thr-148) on Twist1 3D structure (green) shown dimerized with another bHLH protein (gray). Note that the phosphorylation sites (yellow stars) are within the loop and accessible on the exterior of the dimer. WT Twist1 or phospho-mutants were transfected in HEK293T cells (C, panels i and ii) or EOC cells clone R182 (D, panels I and ii) as indicated; the levels of Twist1 protein were determined by Western blotting analysis; and the resulting bands were quantified by densitometry. Densitometry readings are reported as fold changes compared with empty vector control. *, p = 0.0126; **, p = 0.0021. E, WT Twist1 (panel i), phospho-deficient on 144 (144A, panel ii), or phosho-mimic on 144 (144D, panels iii) were transfected in HEK293T cells and treated with cyclohexamide (CHX, 20 μg/ml). The levels of Twist1 protein were determined by Western blotting, and the resulting bands were quantified by densitometry; densitometry readings are reported as fold changes compared with empty vector control. **, p = 0.0096. GAPDH was used as loading control.