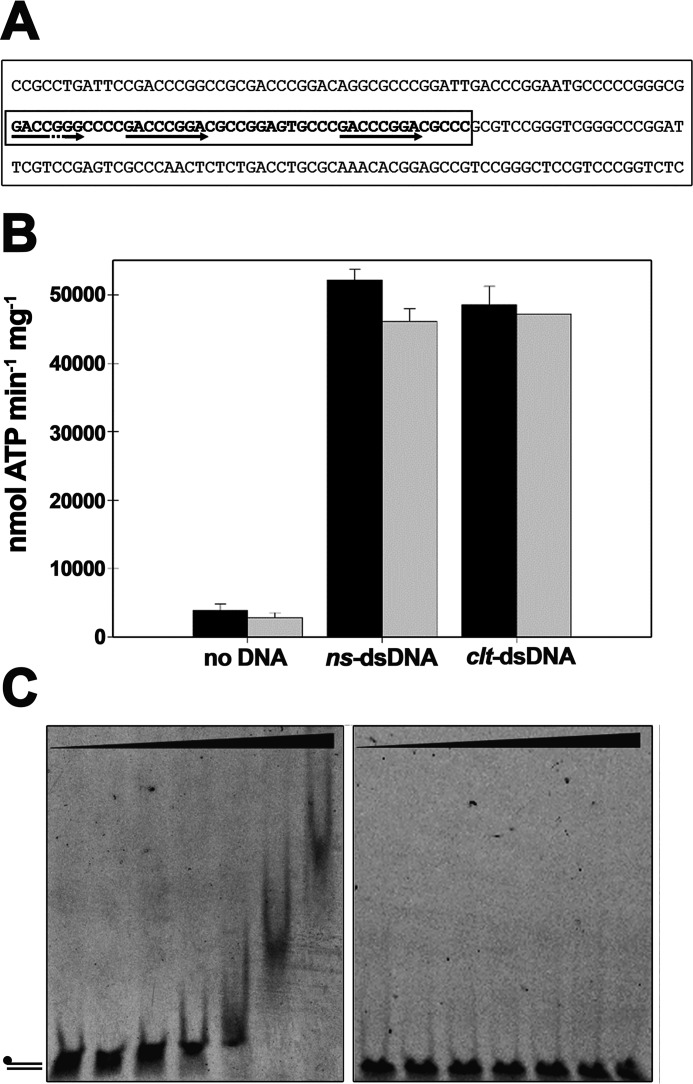
Figure 2.
TraB does not show sequence-specific ATPase stimulation. A, nucleotide sequence of the TraB-binding region of plasmid pSVH1. 45-base clt sequence is within a box, where the two perfect and one imperfect TRS sequences are marked with arrows. B, the ATP turnover of TraBs (black) and TraBsΔγ (gray) was measured in the absence of any DNA substrate, in the presence of nonspecific dsDNA (ns-dsDNA), formed by the annealing of 45-mer nucleotides A and B from Table 1, and in the presence of dsDNA with the clt sequence (clt-dsDNA), formed by the annealing of 45-mer nucleotides C and D from Table 1. Reactions contained either TraBs or TraBsΔγ (50 nm), 5 mm ATP, and 260 nm DNA substrate. Data represent the average of three different experiments. C, gel mobility shift assays. dsDNA with the clt sequence (5 nm) was formed as above, but with 5′-fluorescein–labeled oligonucleotide C (left panel). The mobility shift with nonspecific DNA (oligonucleotides A and B) is shown on the right panel. DNA was incubated with increasing concentrations of TraBs or TraBsΔγ proteins at 0, 0.05, 0.2, 0.5, 1, 2, and 5 μm, as described under “Experimental procedures.” Error bars: S.D.