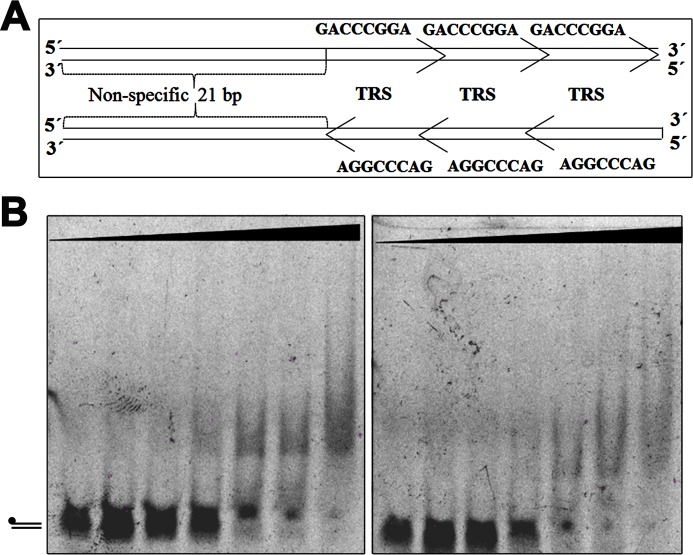
Figure 3.
TraBs does not require a specific orientation of the TRS motifs for DNA binding. A, schematic representation of the two dsDNA substrates used in the assay (3 × TRS, and 3 × reverse TRS). B, native EMSAs using overlapping TRS sequences in the two different orientations. Substrates (dsDNA) were formed by the annealing of oligonucleotides E to F (left panel) and G to H (right panel), respectively. DNA samples (5 nm) were incubated with increasing concentrations of TraBs protein at 0, 0.05, 0.2, 0.5, 1, 2, and 5 μm, as described under “Experimental procedures.”