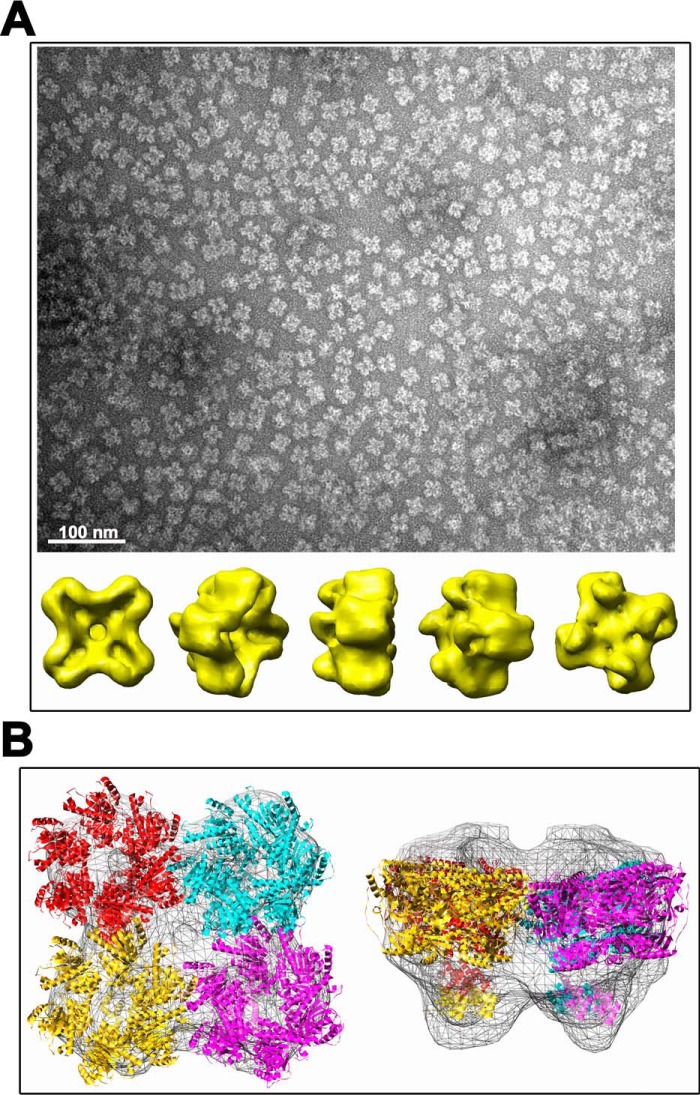
Figure 6.
3D reconstruction of TraBs oligomer. A, TraBs from peak 1 was negatively stained with uranyl acetate and analyzed by EM. Particles were selected, aligned, and classified by maximum likelihood methods. A 3D reconstruction was obtained by projection matching, with imposed C4 symmetry in the final iterations. Scale bar represents 100 nm. B, fitting of TraBs in the EM maps. An atomic model of TraBs was generated by using atomic coordinates from FtsK of P. aeruginosa as template (2iuu.pdb and 2ve9.pdb coordinates for the motor domain and the γ-domain, respectively). The atomic coordinates were docked into the EM maps using SITUS package software.