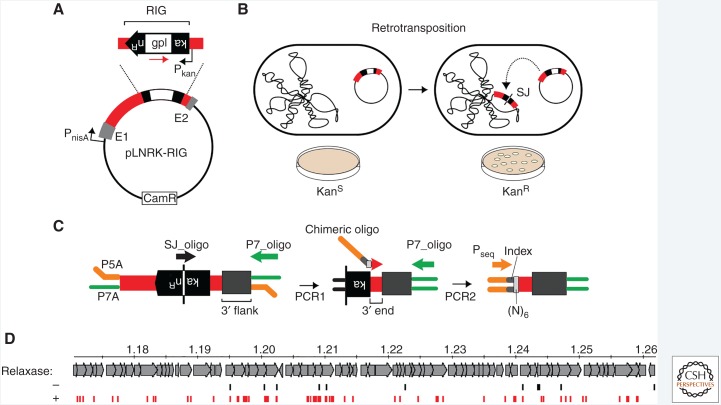
Figure 3.
RIG-seq for high-throughput retrotransposon profiling. (A) RIG assay. The intron donor plasmid pLNRK-RIG carries the Ll.LtrB intron (red) flanked by exons (gray) under a nisin-inducible promoter (PnisA). The intron is interrupted by a retromobility indicator gene (RIG) with the kanR gene inserted in the anti-transcriptional orientation to the intron and carrying its own promoter (Pkan). The kanR gene is interrupted by a self-splicing group I intron (gpI) in the same orientation as Ll.LtrB (red arrow). (B) Reconstitution of the kanR gene with formation of a characteristic splice junction (SJ) sequence and kanamycin resistance is possible only through reverse transcription of an RNA intermediate that lost the group I intron during retrotransposition. Wavy line to the left is chromosomal DNA, whereas plasmid to the right is the RIG donor. (C) RIG-seq amplification scheme. High-throughput targeted sequencing of insertion loci was based on the generation of the SJ sequence of the kanR gene during retrotransposition. After ligation of Illumina-specific adapters, P5A and P7A, two tandem polymerase chain reactions (PCRs) were used to amplify flanks of retrotransposition events in Illumina libraries: A specific SJ primer (SJ_oligo) with the P7_oligo is used in the first PCR; then a library-specific primer, the chimeric oligo, carrying library-specific indices, is used with the P7_oligo in the second PCR. Pseq is the sequencing primer. (D) Density of intron-insertions in part of the Lactococcus lactis chromosome is shown, in which numbers correspond to mbp from the origin of replication, in the presence (+, red) or absence (−, black) of relaxase. (Adapted from Novikova et al. 2014.)